Blood Proteomics Challenges & Solutions: Driving Low-Abundance Protein Discovery
The blood proteome is an invaluable resource in pharmaceutical research and clinical investigations. As an easily accessible biofluid, blood contains a vast array of proteins that mirror the physiological and pathological state of the body. This makes blood proteomics a powerful tool for early disease diagnosis, prognosis assessment, and drug discovery. With the evolution of high-resolution analytical platforms, comprehensive profiling of the plasma proteome is enabling the identification of novel biomarkers and therapeutic targets across oncology, cardiovascular diseases, metabolic disorders, and neurological conditions. Of particular interest are low-abundance proteins, which often play critical roles in signaling and regulation. Thus, in-depth blood proteomic analysis has become essential for uncovering new molecular mechanisms and supporting precision medicine.
Blood Proteome Complexity and Analytical Challenges
Among human proteomes, blood plasma stands out for its extraordinary complexity. Plasma contains thousands of proteins spanning a dynamic range exceeding 10 orders of magnitude. The top 10 most abundant proteins (such as albumin and immunoglobulins) account for roughly 90% of the total protein mass, while a myriad of lower-abundance proteins exist at concentrations that can be billions of times lower. This vast disparity poses a significant hurdle for mass spectrometry-based proteomics, where high-abundance proteins tend to dominate the MS signal, effectively masking the detection of low-abundance proteins. Even with modern mass spectrometers reaching femtomole to attomole sensitivities, direct analysis of untreated plasma often fails to capture proteins present at trace levels. Moreover, many low-abundance signaling molecules may be bound to carrier proteins like albumin, further complicating their detection. Together, these factors render deep proteomic profiling of blood highly challenging, particularly when the aim is to discover novel disease biomarkers or subtle therapeutic effects.
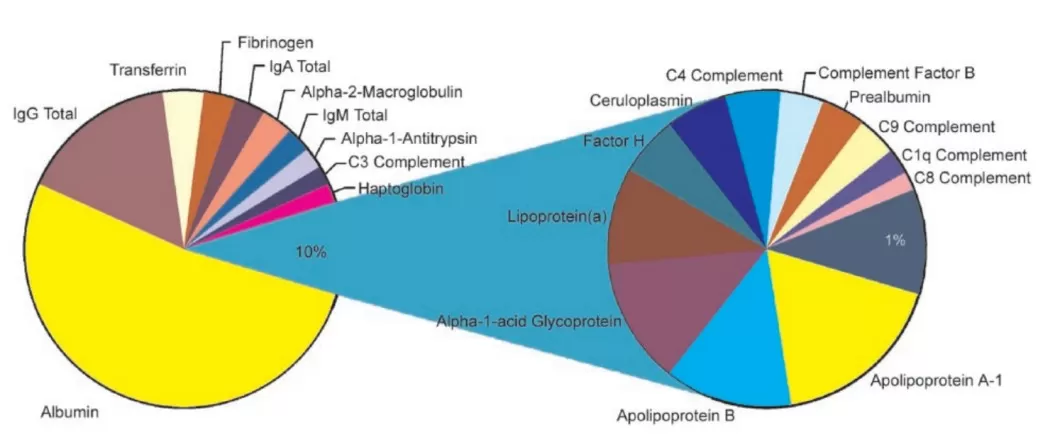
The relative contribution of proteins within plasma (Tirumalai et al., 2003)
Why Low-Abundance Proteins Matter in Drug and Clinical Research
While low-abundance proteins make up a tiny fraction of the plasma proteome by mass, they frequently carry disproportionate biological significance. Many cytokines, hormones, and growth factors exist at picogram to nanogram per milliliter levels but are pivotal in regulating immune responses, cell growth, and tissue homeostasis. Deviations in their concentrations often serve as early indicators of disease onset or progression. Likewise, proteins leaking into the bloodstream from specific tissues due to injury or pathology can act as sensitive early warning markers. For pharmaceutical R&D, these low-abundance proteins frequently represent promising novel drug targets or mechanistic biomarkers. A comprehensive capture of such low-abundance proteomic changes greatly enhances the probability of identifying meaningful disease signatures or validating therapeutic effects. Therefore, robust strategies to uncover these elusive molecules are indispensable for advancing both biomarker-driven clinical studies and innovative drug development.
Current Landscape of Blood Proteomics Technologies
Recent advances have brought non-mass-spectrometry technologies, such as Olink’s proximity extension assays (PEA) and SomaLogic’s SomaScan aptamer platform, to the forefront of blood proteomics. These approaches utilize antibody- or aptamer-based binding, matching each target protein with a specific capture reagent. This design enables exceptional sensitivity and an extremely wide dynamic range, making them highly effective for quantifying low-abundance proteins within targeted studies. As a result, they have become popular choices for large-scale investigations focused on predefined biomarker panels, including expansive clinical cohorts and epidemiological research. However, because they rely on predefined panels and molecular recognition events, they can face specificity challenges and are inherently limited to proteins for which high-quality antibody or aptamer pairs exist.
In contrast, mass spectrometry-based proteomics remains indispensable for blood studies thanks to its unparalleled flexibility and cost-effectiveness. Unlike panel-based technologies that are confined to predefined targets, mass spectrometry enables broad, unbiased exploration of thousands of proteins within a single analysis. This makes it highly adaptable—equally suited for large-scale cohort investigations and for small exploratory studies where biomarkers and pathways have yet to be defined. In addition, tailored workflows allow researchers to uncover critical regulatory proteins that might otherwise go undetected. As a result, while affinity-based platforms like Olink and SomaScan offer exceptional targeted capabilities, mass spectrometry remains the practical, scalable backbone of blood proteomics, supporting a wide range of pharmaceutical and clinical research needs.
Proteomics Platform Showdown: MS-DIA vs. Olink vs. SomaScan
Limitations of Traditional MS-based Blood Proteomics Workflows
Traditional proteomic workflows typically involve direct digestion and LC-MS/MS analysis of plasma or serum samples. However, due to the extreme dynamic range and overwhelming presence of high-abundance proteins, such approaches usually yield only a limited snapshot of the proteome, predominantly featuring medium-to-high abundance proteins. As a result, numerous biologically meaningful low-abundance proteins go undetected, which can severely limit biomarker discovery or mechanistic insights. Furthermore, conventional methods often suffer from reproducibility issues, given the manual, multi-step sample preparation processes prone to variability. This poses additional concerns for large-scale pharmaceutical or clinical studies that require robust, high-throughput and reproducible analyses. Without specialized preprocessing, mass spectrometry analyses are largely confined to the most dominant proteins, missing out on potentially game-changing low-abundance signals.
Essential Strategies for Low-Abundance Protein Enrichment
Given the inherent challenges of plasma proteomics and the immense value of low-abundance proteins, implementing low-abundance protein enrichment strategies has become critical. These approaches aim to reduce sample complexity and compress the dynamic range prior to MS analysis, thereby enhancing the detectability of rare proteins.
- Immunodepletion of high-abundance proteins: One widely adopted strategy is the targeted removal of predominant plasma proteins using immunoaffinity techniques. Commercial multi-affinity removal columns (e.g., MARS) can simultaneously deplete the top 7–14 most abundant proteins, eliminating up to 97–99% of total protein mass. This effectively enriches the remaining proteome for lower-abundance species and has been shown to significantly expand protein identification coverage. However, caution is warranted as proteins bound to the depleted carriers (like albumin) may also be unintentionally removed.
- Specific enrichment of low-abundance proteins: Instead of depleting dominant proteins, another approach focuses on selectively capturing low-abundance proteins. Technologies like ProteoMiner leverage hexapeptide ligand libraries on beads to normalize protein concentrations: high-abundance proteins saturate their ligands and wash off, while low-abundance proteins are retained, enriching their representation in the final sample. Similarly, advanced nanoparticle systems functionalized with bait molecules can simultaneously exclude major plasma proteins and bind diverse low-abundance targets, dramatically increasing their detectability by mass spectrometry.
- Multidimensional fractionation: Beyond enrichment, multi-step chromatographic separations (e.g., strong cation exchange, high/low pH reversed-phase fractionation) help reduce sample complexity by dividing peptides into orthogonal fractions for successive MS analysis. Such multidimensional workflows are often combined with immunodepletion or ligand-based enrichment to maximize proteome coverage, particularly for challenging samples like plasma.
Together, these strategies are pivotal for expanding the depth and breadth of blood proteomics, enabling researchers to capture biologically and clinically relevant low-abundance proteins that would otherwise remain hidden.
Internal data spotlight: Enrichment Boosts Detection Depth
Our in-house studies have quantitatively demonstrated the profound benefits of low-abundance protein enrichment in blood proteomics. In direct workflows without any enrichment, typical plasma LC-MS/MS analyses identified only a few hundred proteins per sample, mostly dominated by high-abundance categories. However, upon incorporating optimized low-abundance enrichment protocols, the number of identified proteins per sample surged to well over a thousand—often reaching 3000–5000 proteins—representing a multi-fold increase. Importantly, many of these newly detected proteins were cytokines, tissue-specific markers, or signaling molecules absent in the unenriched datasets. Functional pathway analyses also revealed a much broader spectrum of biological processes in the enriched datasets, including inflammation, cell communication, and metabolic regulation, whereas unenriched profiles remained narrowly centered around coagulation and complement pathways. This stark contrast underscores how strategic enrichment fundamentally transforms blood proteomics, enhancing both analytical depth and biological interpretability.
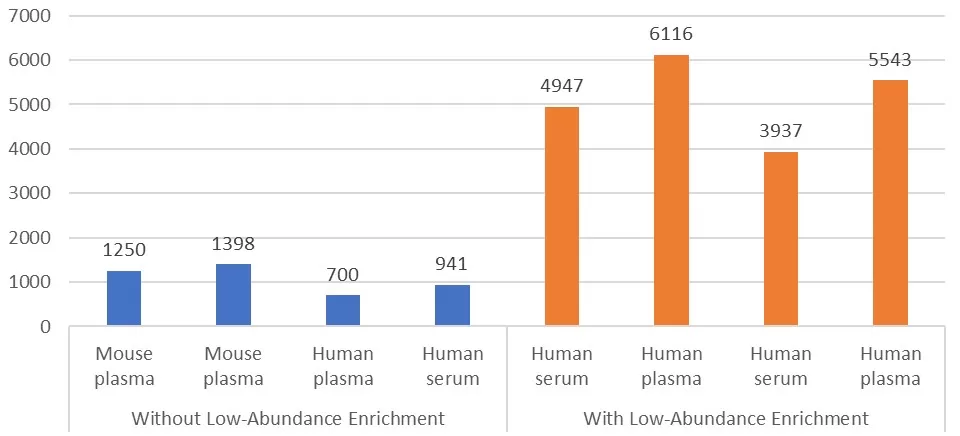
Impact of Low-Abundance Protein Enrichment on Blood Proteome Coverage
Why Choose Us: Leading Platforms & Full-Stack Proteomics Services
As a specialized multi-omics service provider, we offer industry-leading proteomics services tailored for pharmaceutical and clinical research partners. Our strengths include:
State-of-the-art mass spectrometry platforms
We operate cutting-edge Bruker timsTOF instruments featuring PASEF and dual TIMS technologies. This advanced infrastructure ensures exceptional sensitivity, wide linear dynamic range, and high reproducibility across all quantitative proteomics workflows.
Expertise in quantitative proteomics technologies
We specialize in label-free quantitative proteomics, offering robust DIA analyses for in-depth discovery studies. For challenging projects, we provide dedicated solutions such as low-input proteomics for limited sample amounts and specialized low-abundance enrichment workflows tailored for blood proteomics. Combined with advanced data processing pipelines using tools like Spectronaut and MaxQuant, we ensure precise, reproducible quantitation aligned with your pharmaceutical or clinical research goals.
Custom method development and extensive R&D experience
Our scientific team combines expertise in molecular biology, analytical chemistry, and bioinformatics, allowing us to continuously refine enrichment and fractionation protocols specifically for challenging matrices like plasma. We regularly innovate to push the boundaries of low-abundance protein detection, offering truly customized solutions.
Reliable project delivery and technical support
We adhere to rigorous quality control at every step, from sample handling and instrument calibration to data analysis. Each project is delivered with detailed reports covering experimental workflows, QC metrics, protein identifications, quantitative comparisons, and biological pathway insights. Our dedicated scientists also provide one-on-one support to help interpret results in a pharmaceutical or clinical context.
Advancing Drug & Clinical Research with Blood Proteomics
In summary, although blood proteomics presents significant technical hurdles due to its extreme dynamic range and dominant high-abundance proteins, implementing low-abundance protein enrichment and leveraging advanced quantitative proteomics technologies have transformed what is achievable. These breakthroughs open new possibilities for discovering subtle yet critical biomarkers and therapeutic targets. By partnering with us, you gain access to sophisticated workflows, robust data quality, and actionable biological insights that accelerate drug development pipelines and clinical translational research. Contact us to learn how our expertise and mass spectrometry platforms can empower your next proteomics project.
Reference
1. Millioni, Renato et al. “High abundance proteins depletion vs low abundance proteins enrichment: comparison of methods to reduce the plasma proteome complexity.” PloS one vol. 6,5 e19603. 4 May. 2011, doi:10.1371/journal.pone.0019603
2. Tirumalai, Radhakrishna S et al. “Characterization of the low molecular weight human serum proteome.” Molecular & cellular proteomics : MCP vol. 2,10 (2003): 1096-103. doi:10.1074/mcp.M300031-MCP200
3. Tu, Chengjian et al. “Depletion of abundant plasma proteins and limitations of plasma proteomics.” Journal of proteome research vol. 9,10 (2010): 4982-91. doi:10.1021/pr100646w
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


