Proteome + PTM Analysis
Proteome + PTM Analysis
Comprehensive Proteome and PTM Profiling
MetwareBio’s Proteome + PTM Combined Analysis Service offers a unified solution to address this need. Leveraging cutting-edge 4D LC-MS/MS technology and optimized enrichment protocols, the service enables high-resolution quantification of total proteomes and key PTMs through parallel experimental workflows, followed by integrated data analysis to provide a multidimensional view of protein regulation. This approach empowers researchers to dissect signaling pathways, uncover disease mechanisms, and identify novel therapeutic targets with greater precision and biological relevance.
Why Choose MetwareBio for Proteomics and PTM Analysis
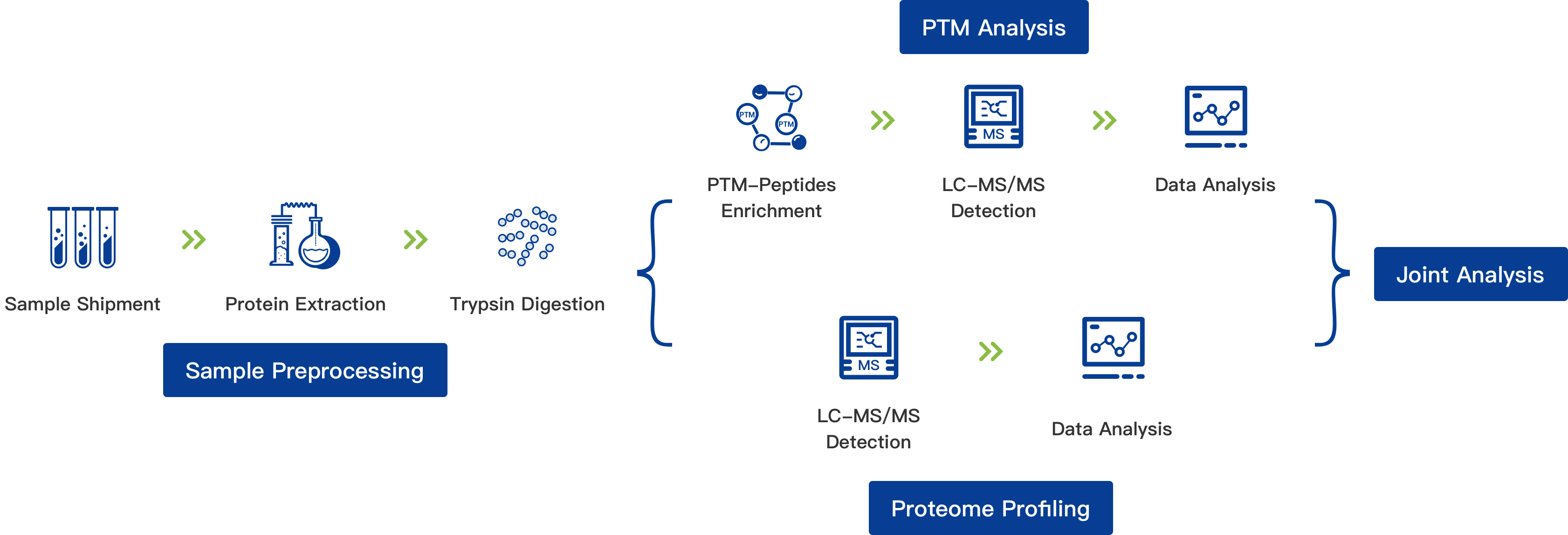
Project Workflow for Proteome + PTM Analysis
Deliverables for Integrated Proteomics and PTM Analysis
Project Experience of Integrated Proteomics and PTM Analysis
Number of phosphorylation sites identified across various medical and plant samples in phosphoproteomics projects
Number of proteins identified from various medical and plant samples in DIA Projects
Applications of Integrated Proteomics and PTM Analysis
Protein expression and post-translational modifications (PTMs) jointly regulate critical signaling pathways in diseases such as cancer, cardiovascular disorders, and neurodegeneration. Integrative proteomics and PTM analysis enables simultaneous profiling of protein abundance and functional states, facilitating the identification of dysregulated pathways, therapeutic targets, and predictive biomarkers. This comprehensive approach enhances mechanistic studies in oncology, immunology, and precision medicine.
In model organisms like mice, rats, and zebrafish, protein expression and PTMs orchestrate key biological events including embryonic development, neuronal plasticity, and immune homeostasis. Combined proteome and PTM analysis supports in vivo functional studies by revealing dynamic regulation of proteins at both the expression and modification levels. This dual-layer insight aids in gene function validation, phenotype correlation, and preclinical model development.
In bacteria, fungi, and viruses, PTMs such as phosphorylation and acetylation modulate virulence, metabolism, and antibiotic resistance. Meanwhile, hosts employ complex proteomic and PTM responses during pathogen challenge. Integrated analysis of host and microbial protein regulation provides a powerful tool for dissecting infection mechanisms, immune evasion strategies, and pathogen-induced signaling alterations, supporting vaccine research and anti-infective drug discovery.
Plants rely on tightly regulated protein networks and PTMs to respond to environmental stimuli, such as drought, temperature stress, and pathogen invasion. Proteome + PTM joint profiling enables detailed investigation of signaling cascades involved in growth, hormone signaling, and stress adaptation. This strategy empowers research in plant functional genomics, crop improvement, and stress resistance engineering.
Sample Requirements for Proteomics Services
| Sample Type | Samples | DIA/DDA Proteomics /PRM Targeted Proteomics |
Phosphorylation Proteomics | Ubiquitin Proteomics/ Acetyl-Proteomics |
|||
| Recommended Sample Size | Minimum Sample Size | Recommended Sample Size | Minimum Sample Size | Recommended Sample Size | Minimum Sample Size | ||
| Human/Animal Tissue | Normal tissues (heart, liver, spleen, lungs, intestines, kidneys, red bone marrow etc.) | 20mg | 5mg | 30mg | 15mg | 100mg | 50mg |
| Yellow bone marrow | 50mg | 20mg | 50mg | 30mg | 200mg | 100mg | |
| Bone, shells, eggshell | 1g | 500mg | / | / | / | / | |
| Hair | 500mg | 200mg | / | / | / | / | |
| Fatty tissue, Skin, synovial membrane | 200mg | 100mg | / | / | / | / | |
| Liquid Samples | Serum/Plasma | 10μL | 1μL | 100μL | 20μL | / | / |
| Serum/Plasma (remove high abundance proteins) | 100μL | 50μL | / | / | / | / | |
| Cerebrospinal fluid, Joint fluid, Lymph fluid, ascites | 200μL | 100μL | / | / | / | / | |
| Amniotic fluid | 200μL | 100μL | 200μL | 100μL | 600μL | 300μL | |
| Milk | 50μL | 10μL | 200μL | 100μL | 600μL | 300μL | |
| Aqueous humor, Vitreous body, Tear fluid | 200μL | 100μL | / | / | / | / | |
| Tear fluid | 200μL | 100μL | / | / | / | / | |
| Urine, alveolar lavage fluid (BALF) | 5mL | 2mL | / | / | / | / | |
| Cellular supernatant | 25mL | 10mL | / | / | / | / | |
| Fermentation broth | 10mL | 5mL | / | / | / | / | |
| Lipid droplet | 300μL | 200μL | / | / | / | / | |
| Saliva (mammals) | 200μL | 100μL | / | / | / | / | |
| Cells | Primary Cells | 3×10^6 | 1×10^6 | 2×10^7 | 1×10^7 | 2×10^7 | / |
| Transmissible cells | 1×10^6 | 5×10^5 | 1×10^7 | 5×10^6 | 2×10^7 | / | |
| Sperm, Platelets | 1×10^7 | 5×10^6 | 1×10^8 | 5×10^7 | 4×10^8 | 2×10^8 | |
| Plant Tissue | Young tissue (young leaf, seedling, petal, etc.) | 50mg | 20mg | 200mg | 100mg | 1g | 500mg |
| Mature tissue (root, stem, fruit, pericarp, etc.) | 500mg | 200mg | 500mg | 300mg | 2g | 1g | |
| Bark, root, fruit | 1g | 500mg | 3g | 1.5g | 5g | 3g | |
| Pollen | 40mg | 15mg | / | / | / | / | |
| Microorganisms | Bacteria | 100mg | 50mg | 300mg | 150mg | 500mg | 200mg |
| Fungi | 200mg | 100mg | 300mg | 150mg | 1g | 500mg | |
| Special samples | Exosome (sediment) | 25μL | 15μL | / | / | / | / |
| Protein | Protein | 100μg | 50μg | 1mg | 500μg | 5mg | 3mg |
Biological duplicates: A minimum of 3 replicates is required; 3-6 replicates for animal samples; 6-10 for clinical samples.
FAQ
Combining total proteome quantification with PTM profiling provides a more comprehensive understanding of protein regulation. While proteomics reveals protein abundance, PTM analysis uncovers dynamic functional modifications such as phosphorylation or ubiquitination, enabling deeper insights into biological signaling and regulatory mechanisms.
Our platform currently supports high-resolution quantitative analysis of key PTMs including phosphorylation, acetylation, ubiquitination, succinylation, and others. Customized enrichment strategies are used for each PTM type to ensure specificity and sensitivity.
Total protein and PTM samples are processed in parallel workflows. Protein extracts are digested into peptides, followed by enrichment of modified peptides (e.g., using phosphopeptide or ubiquitin remnant affinity methods) before LC-MS/MS analysis. This approach ensures optimal detection of both total proteins and specific PTMs.
All analyses are performed using state-of-the-art 4D label-free LC-MS/MS platforms such as timsTOF systems, which provide high sensitivity, deep proteome coverage, and accurate quantification of both total proteins and modified peptides.
Currently, our service is based on label-free quantification, which offers excellent detection sensitivity and broad proteome coverage at a lower cost. With advancements in mass spectrometry technology, label-free proteomics has become a reliable and efficient approach for both total protein and PTM quantification across multiple biological conditions.
Our bioinformatics team performs joint analysis of protein expression and PTM datasets, using pathway enrichment, correlation analysis, clustering, and regulatory network mapping. This integrative approach links changes in protein levels with functional modification patterns, enhancing biological interpretation.
Typically, 150–300 µg of total protein is required per sample for combined analysis, depending on the PTM type and enrichment strategy. We accept a range of biological materials, including cells, tissues, serum/plasma, and model organisms. Please contact us for specific sample preparation guidelines.
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


