Untargeted Metabolomics
Untargeted Metabolomics
What is Untargeted Metabolomics?
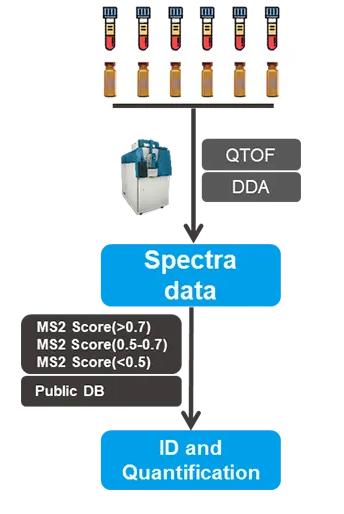
Why Choose MetwareBio’s Untargeted Metabolomics Service?
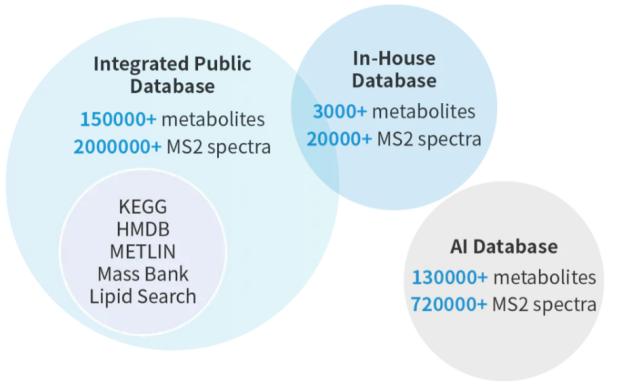
List of Metabolites
MetwareBio’s untargeted metabolomics platform offers broad and in-depth coverage of metabolites involved in key biological pathways. Detectable compound classes include amino acids, carbohydrates, organic acids, nucleotides, lipids, amines, alcohols, ketones, aldehydes, steroids, bile acids, vitamins, and various secondary metabolites. These metabolites span critical pathways such as energy metabolism, amino acid metabolism, nucleotide biosynthesis, lipid metabolism, and redox balance, enabling comprehensive insights into cellular function and systemic metabolic regulation.
In-house biomedical database
| Category | Quantity | Representative substance |
| Amino acids and their derivatives | 600+ | glycine, L-threonine, L-arginine, N-acetyl-L-alanine |
| Organic acids and their derivatives | 400+ | 3-hydroxybutyric acid, adipic acid, hippuric acid, kynurenine |
| Nucleotides and their derivatives | 200+ | adenine, 5'-Adenine Nucleotide, guanine, 2'-Deoxycytidine |
| Carbohydrates and their derivatives | 100+ | D-glucose, glucosamine, D-fructose 6-phosphate |
| Lipids | 500+ | O-acetylcarnitine, γ-linolenic acid, lysophosphatidylcholine 22:4 |
| Benzene and its derivatives | 500+ | benzoic acid, 3,4-dimethoxyphenylacetic acid, 4-hydroxybenzoic acid |
| Coenzymes and vitamins | 60+ | folic acid, pantothenic acid, vitamin D3 |
| Alcohols, Amines | 150+ | dopamine, histamine, DL-1-amino-2-propanol |
| Aldehydes, Ketones, Esters | 120+ | furfural, ethyl butyrate, α-pentyl cinnamaldehyde |
| Heterocyclic compound | 200+ | pyridoxal, biopterin, indole-3-acetic acid |
| Bile acids | 40+ | glycocholic acid, deoxycholic acid, taurolithocholic acid |
| Hormones and hormone-related substances | 100+ | juvenile hormone 3, epinephrine, 3,3'-diiodo-L-thyroxine |
| Tryptamines, Cholines, Pigments | 15+ | serotonin, bilirubin (E-E), urobilin |
| Others | 50+ | astaxanthin, hydroxyurea |
| Total | 3000+ | |


Quantification

Deliverables of Untargeted Metabolomics Service
Demo Report Of Untargeted Metabolomics
Experience in Untargeted Metabolomics Service
Number of metabolites identified from various sample types using MetwareBio's untargeted metabolomics approach
Applications of Untargeted Metabolomics Service
Untargeted metabolomics plays a crucial role in exploring disease mechanisms and identifying potential biomarkers. By profiling global metabolic changes in patient samples, researchers can uncover metabolic signatures associated with cancer, metabolic disorders, neurodegenerative diseases, and more. This approach supports early diagnosis, patient stratification, and therapeutic monitoring in clinical and translational research.
In pharmacology research, untargeted metabolomics enables comprehensive evaluation of drug-induced metabolic changes. It helps assess drug efficacy, toxicity, and off-target effects by capturing metabolic shifts in blood, tissue, or urine samples. This approach is widely used in preclinical studies to support drug mechanism elucidation and safety evaluation.
Untargeted metabolomics is widely applied in animal models and livestock research to study physiological responses to diet, environment, or disease. By analyzing metabolic profiles in serum, tissue, feces, and milk, researchers can monitor growth, immunity, and health status. This helps optimize animal nutrition strategies and improve welfare in veterinary and agricultural settings.
This approach is a powerful tool for studying microbial metabolism and host-microbiome interactions. It allows researchers to track microbial-derived metabolites, analyze gut microbial activity, and explore how microbiota influence host physiology. Applications span from probiotic research and microbiome-targeted therapies to understanding microbial roles in inflammation and metabolic diseases.
Case Study of Untargeted Metabolomics Service
Untargeted Metabolomics Reveals Distinct Metabolic Responses to Dual Nutrient Delivery in the Small Intestine
In a Cell publication titled “A two-front nutrient supply environment fuels small intestinal physiology through differential regulation of nutrient absorption”, researchers examined how luminal and vascular nutrient inputs shape the metabolic environment of the small intestine. Using a mouse model under sham feeding, total parenteral nutrition (TPN), and starvation conditions, gut interstitial fluid (GIF) was collected for untargeted metabolomics analysis performed by MetwareBio. The results showed that enteral nutrient input enriched lipid-related and steroid biosynthesis metabolites, while serosal delivery led to higher levels of carbohydrates and organic acids. Importantly, many of these metabolites were microbiota-derived, suggesting a key role of host–microbiome interactions in shaping intestinal physiology.
This study demonstrates the power of untargeted metabolomics in dissecting nutrient-specific metabolic regulation. MetwareBio’s platform enabled the unbiased profiling of extracellular metabolite dynamics, revealing distinct biosynthetic pathways engaged by different feeding routes. This approach offers valuable insights for research in nutrition, gut health, and host–microbe interactions.
GIF extraction and detection in the small intestine (Zhang et al., 2024)
Sample Requirements of Untargeted Metabolomics Service
| Sample Type | Sample | Recommended sample size | Minimum sample size | Biological replication |
| Liquid | Plasma, serum, hemolymph, milk, egg white | 100μL | 20μL | human>30 |
| animal>6 | ||||
| Cerebrospinal fluid, tear fluid, interstitial fluid, uterine fluid, pancreatic, fluid and bile, pleural effusion, follicular fluid, corpse fluid | 100μL | 20μL | human>30 | |
| animal>6 | ||||
| seminal plasma, amniotic fluid, prostate fluid, rumen fluid, respiratory, condensate, gastric lavage fluid, alveolar lavage fluid, urine, sweat, saliva, sputum | 500μL | 100μL | human>30 | |
| animal>6 | ||||
| Tissue | Animal tissue, placenta, thrombus, fish skin, mycelium, nematode | 100mg | 20mg | human>30 |
| animal>6 | ||||
| whole body, aircraft (wings). pupae | 500mg | 20mg | human>30 | |
| animal>6 | ||||
| Zebrafish organs, insect organs | 20 | 10 | animal>6 | |
| Cell | Cultured cells | 2*10^6 | 1*10^6 | human>30 |
| animal>6 | ||||
| Escherichia coli and other microorganisms | 1*10^10 | 5*10^8 | animal>6 | |
| Feces | Feces, ilntestinal contents | 200 mg (wet weight) | 50 mg (wet weight) | human>30 |
| animal>6 |
FAQ on Untargeted Metabolomics
Untargeted metabolomics captures a broad range of metabolites in a sample without prior selection, making it suitable for discovery-driven research and hypothesis generation. Targeted metabolomics, by contrast, focuses on quantifying a defined set of known metabolites, often with absolute quantification and greater sensitivity for specific compounds.
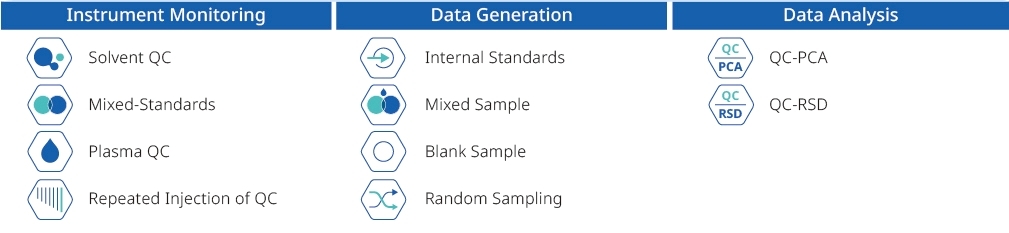
A robust experimental design should include adequate biological replicates, matched controls, and consistent sample handling to reduce variability. Incorporating quality control (QC) samples, such as pooled QC and reference standards, is essential for monitoring data consistency. We also recommend randomizing sample processing and acquisition to minimize batch effects.
We use a combination of QC-based normalization, signal drift correction, and statistical modeling to minimize batch effects. System suitability and pooled QC samples are regularly injected to monitor instrument stability, and multivariate analyses like PCA are applied to assess data quality across batches.
Yes. Unidentified features—metabolic signals without definitive structural annotation—can still be statistically analyzed and used for pattern recognition, classification, or pathway-level inference (via correlation with known metabolites or network-based enrichment). These features may serve as potential biomarkers pending further validation.
Absolutely. We support multi-omics integration through pathway enrichment tools and correlation-based analysis. Combining metabolomics with transcriptomic or proteomic data allows for deeper insight into biological regulation, linking metabolic phenotypes with gene expression and protein activity.
While untargeted metabolomics is inherently more variable than targeted methods, we apply strict quality control protocols, standardized extraction procedures, and internal normalization strategies to ensure the reproducibility. We routinely evaluate CV distribution across QC samples and offer cross-batch comparability analysis for long-term or multi-phase projects.
Reference
Zhang, J., Tian, R., Liu, J., Yuan, J., Zhang, S., Chi, Z., Yu, W., Yu, Q., Wang, Z., Chen, S., Li, M., Yang, D., Hu, T., Deng, Q., Lu, X., Yang, Y., Zhou, R., Zhang, X., Liu, W., & Wang, D. (2024). A two-front nutrient supply environment fuels small intestinal physiology through differential regulation of nutrient absorption and host defense. Cell, 187(22), 6251–6271.e20.https://doi.org/10.1016/j.cell.2024.08.012
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


