Best Practices for Blood Proteomics Sample Prep: Selecting Anticoagulants and Optimizing Pre-analytical Variables
Blood samples are a cornerstone in proteomics research, offering a rich source of biomarkers for disease progression, drug response, and clinical diagnostics. However, the success of plasma proteomics analysis heavily depends on the proper handling and preparation of these samples. Even minor variations in pre-analytical conditions, such as anticoagulant choice, centrifugation settings, and sample processing methods, can lead to significant differences in proteomic outcomes. In this article, we discuss how to select the right anticoagulant for your plasma proteomics study and why blood sample preparation is key to obtaining reliable, high-quality results.
Why Blood Samples are Crucial for Proteomics Research
Blood is an essential biological fluid in proteomics research, providing a vast array of information crucial for identifying disease biomarkers and understanding drug responses. Its non-invasive collection method and rich biological composition make it a preferred choice for clinical studies. Plasma, as the liquid component of blood, holds significant potential as a source for protein biomarkers associated with disease progression and therapeutic outcomes [1,2]. Recent advancements in mass spectrometry have further fueled interest in plasma proteomics, allowing for deeper insights into the proteins that drive health and disease.
However, obtaining accurate and reliable proteomic data from blood samples requires careful attention to pre-analytical conditions. The choice of anticoagulant, centrifugation settings, and sample storage techniques can all affect protein abundance and data quality [3-5]. In large-scale, multi-center studies, variations in how samples are processed can introduce significant inconsistencies, which in turn can impact the proteomic analysis. The Human Plasma Proteome Project (HPPP) is working to standardize these methodologies to improve the reliability and reproducibility of plasma proteomics [1]. Therefore, optimizing blood sample handling procedures is essential for successful biomarker discovery and validation.
Principles and Applications of Different Blood Collection Methods
Blood sample collection methods mainly involve the choice of different anticoagulants and blood collection tubes. Over the past two decades, EDTA plasma has been used as the preferred sample matrix for human blood proteomics analysis, with serum also being widely used [5]. The most commonly used in routine diagnostics are serum or Li-heparin plasma, while the proteomics community mainly focuses on enhancing the analysis depth of EDTA plasma. Currently, there are five blood collection methods, of which three are plasma collection methods (EDTA plasma, citrate plasma, Li-heparin plasma) and two are serum collection methods (conventional serum, serum with separation gel) [6]. The principles of different methods are as follows:
a) EDTA Anticoagulated Plasma (EDTA-Plasma): EDTA chelates calcium ions, inhibiting blood clotting.
b) Heparin Anticoagulated Plasma (Heparin-Plasma): Lithium heparin enhances the activity of antithrombin III to prevent clotting.
c) Citrate Anticoagulated Plasma (Citrate-Plasma): Sodium citrate chelates calcium ions, also preventing clotting.
d) Serum (Serum / Serum with Separation Gel): Blood naturally clots without anticoagulant, and the liquid portion is separated, with or without a separation gel.
A detailed comparison is shown in the following table:
Comparison Table of of Different Blood Collection Methods
|
Sample Type |
Collection Method |
Core Principle & Key Technical Points |
Key Features |
Typical Application |
|
Plasma |
EDTA Plasma |
Anticoagulation principle: Strong calcium chelation to block clotting |
Retains all coagulation factors and fibrinogen, accurate platelet count |
Preferred for hematology analysis: CBC, blood morphology |
|
Citrate Plasma |
Anticoagulation principle: Weaker calcium chelation than EDTA, strict anticoagulant-to-blood ratio |
Minimal interference with coagulation factors, closely resembles in vivo physiological state |
Gold standard for coagulation function tests: PT, APTT, coagulation factor activity |
|
|
Li-Heparin Plasma |
Anticoagulation principle: Enhances antithrombin activity to inhibit thrombin formation |
Avoids electrolyte interference, may induce platelet aggregation |
Common choice for clinical chemistry: glucose, lipids, liver/kidney functions |
|
|
Serum |
Conventional Serum |
Natural coagulation without anticoagulant, blood separates after coagulation |
Lacks fibrinogen and consumed coagulation factors, stable composition |
Used in serology, immunology: antibody testing, tumor markers, hormones |
|
Serum with Separation Gel |
Physical separation using thixotropic gel between serum and blood cells |
Forms a stable barrier preventing cell-material exchange, highly stable |
Ideal for long-term storage, sample transport, and automation-friendly |
How Blood Sample Preprocessing Affects Plasma Proteomics Results
Impacts of Different Anticoagulants on Plasma Proteomics
Studies indicate that EDTA plasma and serum outperform citrate plasma in terms of the number of proteins identified. When comparing heparin plasma with EDTA plasma and serum using scalable automated proteomics workflows (ASAP), 442 proteins were commonly identified in both EDTA plasma and heparin plasma. However, only around 70% of the proteomes overlapped between these blood matrices, suggesting that heparin plasma data can complement those obtained from EDTA plasma [4].
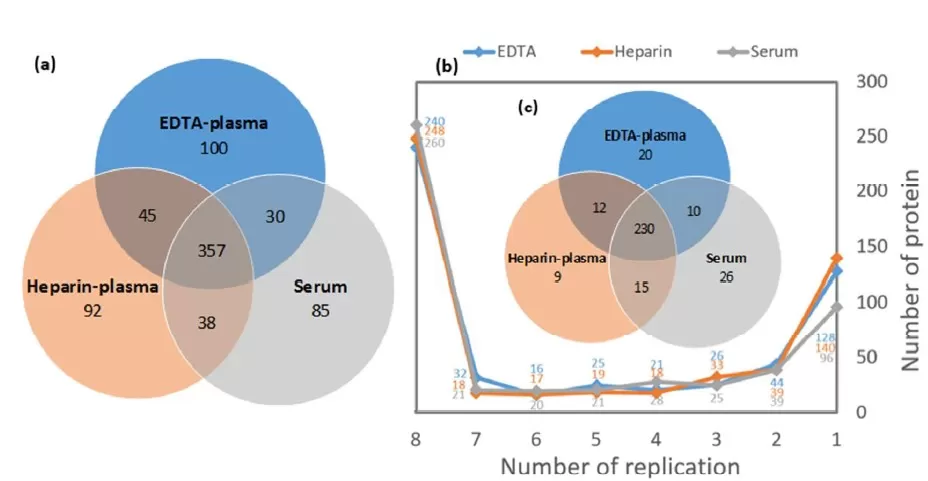
Figure 1: Differences in protein identification across various blood samples.
(a) the protein coverage from commercially matched EDTA plasma, heparin plasma, and serum mixed samples; (b) Reproducibility of identification across 8 replicates, with proteins identified in at least 7 replicates considered consistent identifications [4]
Influences of Pre-analytical Variables on Blood Proteomic Analysis
Pre-analytical variables, such as blood processing and storage, significantly impact biomarker discovery and clinical testing. Delays in centrifugation are particularly problematic, leading to increased proteome variability. Storage temperature and anticoagulant type also play a role, though their effects are comparatively less pronounced [7]. Studies show that delays of less than 6 hours in pre-processing have minimal impact on immune-depleted plasma proteomes, while significant changes occur after 4 days [3].
Specific pre-analytical conditions can significantly alter the plasma proteome. For example, low centrifugal force (1300×g) combined with blood storage at 0°C for 6 hours led to substantial changes in the proteome, with 200 and 148 proteins significantly altered. Storing plasma at 4°C or room temperature for 24 hours also induced changes in protein composition [8].
Effects of Sample Processing Methods on Protein Identification
The method used to prepare samples plays a crucial role in protein identification. Techniques such as strong anion exchange (SAX) magnetic beads, ENRICH-iST kits, and perchloric acid precipitation lead to the highest protein identifications but introduce more variability. Notably, sample preparation methods have a greater impact on the proteome profile than the blood matrix itself [9,10]. Additionally, delayed processing in multi-center studies can significantly alter intracellular proteins, with variations tied to collection site differences, suggesting that inconsistencies in blood processing protocols must be accounted for.
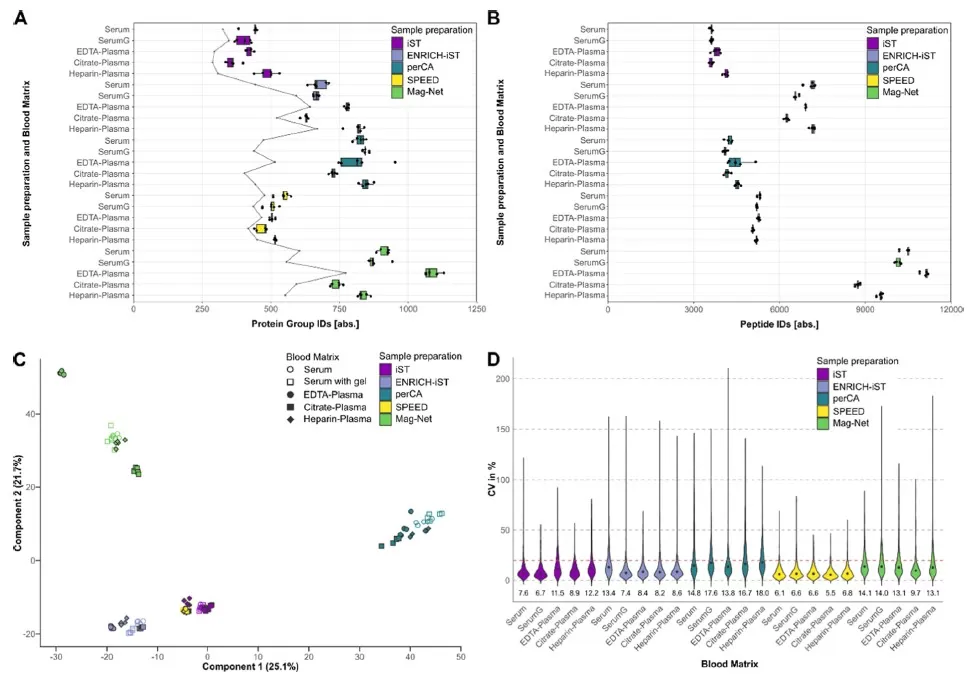
Figure 2: Effects of different blood types and processing methods on proteomics outcomes.
(A) Boxplot representation of proteome identification obtained through five sample preparation methods and five different blood matrix combinations (5 technical replicates). The gray line indicates the number of proteins with a CV value <20% in LFQ levels. (B) Boxplot representation of peptide identifications obtained through five sample preparation methods and five different blood matrix combinations. (C) Principal component analysis of LFQ intensities after missing value imputation and normalization, filtered and log2-transformed. (D) Violin plot based on CV values of proteome LFQ intensities [9].
Comparison of Different Blood Sample Types for Plasma Proteomics
In plasma proteomics, choosing the right blood sample type is crucial for obtaining accurate and reliable results. Each blood sample type—whether EDTA plasma, citrate plasma, Li-heparin plasma, or serum—has its unique advantages and limitations depending on the specific analysis goals. Understanding the differences between these sample types and their impact on proteomic data is essential for optimizing your experimental design and ensuring high-quality proteomics analysis. Below is a comparison of various blood sample types commonly used in plasma proteomics.
Comparison of Blood Sample Types for Plasma Proteomics Analysis
|
Sample Type |
Key Advantages |
Main Disadvantages |
Considerations & Applications |
|
EDTA Plasma |
High proteome integrity: Retains key proteins, closest to physiological state |
High-abundance proteins can obscure low-abundance proteins |
Best for exploratory research aiming to map the complete plasma proteome |
|
Citrate Plasma |
Minimal interference with mass spectrometry, good for downstream analysis |
Anticoagulant may interfere with certain experiments |
Ideal when EDTA plasma is incompatible with laboratory protocols |
|
Heparin Plasma |
May enhance stability of certain proteins by inhibiting proteases |
Potential interference with mass spectrometry or biochemical tests |
Suitable as an alternative to EDTA plasma in certain laboratory setups |
|
Conventional Serum |
Simplified sample processing: No fibrinogen interference |
Coagulation alters the protein composition, leading to potential distortion |
Useful for targeted protein studies where coagulation does not affect stability |
|
Serum with Separation Gel |
Stable, reduces protein degradation, ideal for transport |
Gel may non-specifically adsorb proteins, affecting quantification |
Practical for large-scale studies and samples requiring long-term storage or shipping |
Optimizing Blood Sample Processing for Reliable Plasma Proteomics
The collection and processing of blood samples play a crucial role in ensuring the reliability and quality of plasma proteomics analysis. Variations in sample handling, including the choice of anticoagulant, centrifugation settings, and storage conditions, can significantly affect proteomic results. In this section, we summarize key takeaways and offer practical recommendations for optimizing blood sample processing to achieve consistent, high-quality data for proteomics research.
Select the Right Anticoagulant for Plasma Proteomics
Choosing the appropriate anticoagulant is critical for achieving reliable proteomic results. EDTA plasma and serum are generally preferred for protein identification due to their ability to retain most plasma proteins. EDTA plasma, in particular, is widely recognized as the gold standard for plasma proteomics analysis, offering a comprehensive view of the plasma proteome. However, other anticoagulants such as citrate and heparin plasma may provide complementary data and may be better suited for specific types of analyses, such as coagulation tests or when dealing with anticoagulant-specific limitations in laboratory workflows.
Recommendation:
- For most proteomics studies, EDTA plasma is the preferred choice due to its comprehensive proteome coverage.
- Consider using citrate or heparin plasma for specific studies that require minimal interference with coagulation factors or when working with systems that are not compatible with EDTA.
Minimize Pre-analytical Variability in Plasma Proteomics
Pre-analytical factors such as centrifugation conditions, processing delays, and storage temperatures play a crucial role in the accuracy and reproducibility of plasma proteomics data. Variations in these conditions can introduce inconsistencies in protein recovery, stability, and overall data quality. Delays in processing, particularly between blood collection and centrifugation, can result in protein degradation and significant changes in protein profiles, leading to unreliable results. Standardizing centrifugation protocols, such as using consistent speeds and times across all samples, can help mitigate variability. Similarly, improper storage conditions, such as fluctuating temperatures or multiple freeze-thaw cycles, can cause protein degradation and loss. To preserve protein integrity, plasma samples should be processed and stored promptly, ideally frozen at -80°C in small aliquots to prevent degradation from repeated thawing.
Recommendation:
- Process plasma samples as soon as possible, ideally within 30 minutes of collection, and standardize centrifugation settings (e.g., 1000–2000×g).
- Store plasma at -80°C immediately after processing and avoid repeated freeze-thaw cycles by aliquoting the samples.
- Implement standardized pre-analytical procedures across all samples to minimize variability, especially in multi-center studies.
Implement a Rigorous Quality Control Process for Plasma Proteomics
To ensure the consistency and reliability of proteomics data, implement a robust proteomics quality control (QC) process that includes assessing sample quality at each step of the workflow. This may involve testing for hemolysis, monitoring protein concentration, and assessing sample integrity before proceeding with proteomics analysis.
Recommendation:
- Perform quality checks on samples before starting proteomics analysis, including visual inspection for hemolysis and measuring protein concentration.
- Consider implementing a standardized QC procedure to monitor consistency across all sample batches.
References:
1. Rai AJ, Gelfand CA, Haywood BC, et al. HUPO Plasma Proteome Project specimen collection and handling: towards the standardization of parameters for plasma proteome samples. Proteomics. 2005;5(13):3262-3277. doi:10.1002/pmic.200401245
2. Elliott P, Peakman TC; UK Biobank. The UK Biobank sample handling and storage protocol for the collection, processing and archiving of human blood and urine. Int J Epidemiol. 2008;37(2):234-244. doi:10.1093/ije/dym276
3. Hassis ME, Niles RK, Braten MN, et al. Evaluating the effects of preanalytical variables on the stability of the human plasma proteome. Anal Biochem. 2015;478:14-22. doi:10.1016/j.ab.2015.03.003
4. Lan J, Núñez Galindo A, Doecke J, et al. Systematic Evaluation of the Use of Human Plasma and Serum for Mass-Spectrometry-Based Shotgun Proteomics. J Proteome Res. 2018;17(4):1426-1435. doi:10.1021/acs.jproteome.7b00788
5. Ignjatovic V, Geyer PE, Palaniappan KK, et al. Mass Spectrometry-Based Plasma Proteomics: Considerations from Sample Collection to Achieving Translational Data. J Proteome Res. 2019;18(12):4085-4097. doi:10.1021/acs.jproteome.9b00503
6. McCafferty C, Letunica N, Swaney E, et al. Blood Collection Processing and Handling for Plasma and Serum Proteomics. Methods Mol Biol. 2023;2628:33-40. doi:10.1007/978-1-0716-2978-9_3
7. Halvey P, Farutin V, Koppes L, et al. Variable blood processing procedures contribute to plasma proteomic variability. Clin Proteomics. 2021;18(1):5. Published 2021 Jan 19. doi:10.1186/s12014-021-09311-3
8. Daniels JR, Cao Z, Maisha M, et al. Stability of the Human Plasma Proteome to Pre-analytical Variability as Assessed by an Aptamer-Based Approach. J Proteome Res. 2019;18(10):3661-3670. doi:10.1021/acs.jproteome.9b00320
9. Gronauer TF, Merl-Pham J, von Toerne C, Habler K, Teupser D, Hauck SM. Blood Matrices and Sample Preparation Influence Blood Marker Discovery. J Proteome Res. 2026;25(1):405-417. doi:10.1021/acs.jproteome.5c00836
10. Korff K, Müller-Reif JB, Fichtl D, et al. Pre-analytical drivers of bias in bead-enriched plasma proteomics. EMBO Mol Med. 2025;17(11):3174-3196. doi:10.1038/s44321-025-00309-0
Read more
- Blood Proteomics Challenges & Solutions: Driving Low-Abundance Protein Discovery
- Overcoming Dynamic Range in MS-Based Blood Proteomics: Depletion and Enrichment Methods
- Blood Proteomics: Serum or Plasma – Which Should You Choose?
- Proteomics Quality Control: A Practical Guide to Reliable, Reproducible Data
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


