Blood Proteomics: Serum or Plasma – Which Should You Choose?
Blood proteomics has become a cornerstone for biomarker discovery and translational research, but a critical question often arises: should we use serum or plasma for proteomic analysis? Both serum and plasma are derived from blood and widely used in plasma proteomics and serum proteomics studies, yet their differences can significantly impact results. In this blog, we delve into the serum vs. plasma debate in blood proteomics, comparing their preparation, protein content, and suitability for mass spectrometry-based proteomic biomarker studies. We will see why many experts lean toward using plasma for proteomics (especially for discovery of blood-based biomarkers), backed by recent research findings. The goal is to provide a clear, professional comparison to help you choose the right sample type for your proteomic experiments.
Serum vs. Plasma – How Do They Differ?
Both serum and plasma are fluid components of blood, but they are obtained differently and have distinct compositions. Understanding these differences is crucial for blood proteomics:
Plasma: The liquid portion of blood that remains when clotting is prevented. To prepare plasma, blood is drawn into a tube containing an anticoagulant (such as EDTA, heparin, or citrate) and then centrifuged to remove cells. The result is a straw-colored fluid that contains all the proteins present in blood including clotting factors. In other words, plasma = serum plus the clotting proteins.
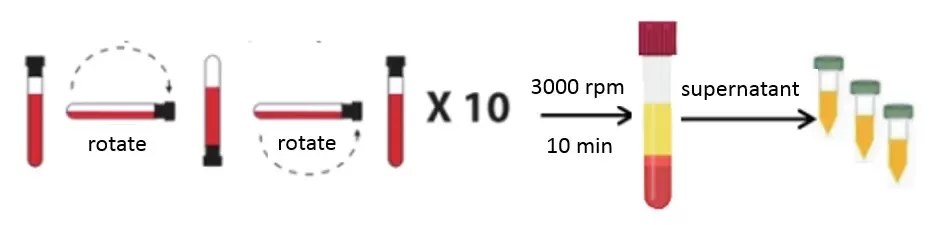
Plasma Preparation Method
Serum: The liquid portion of blood after clotting. To prepare serum, blood is drawn without anticoagulant and allowed to clot at room temperature (typically 30~60 minutes), then centrifuged to remove the clot and blood cells. The remaining clear, yellowish fluid is serum, which contains most blood proteins except those used up in the clotting process.
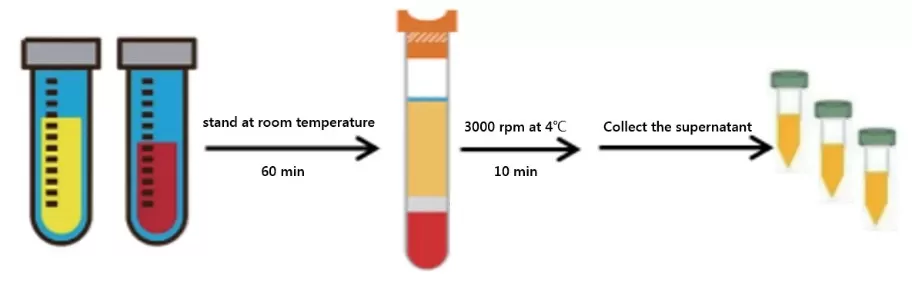
Serum Preparation Method
In simple terms, the key difference can be summed up in one word: “clotting.” Plasma is obtained by preventing clotting (thus retaining clotting proteins in the sample), whereas serum is obtained after allowing the blood to clot (thereby depleting certain clotting-related proteins in the sample). This fundamental distinction leads to several practical differences between the two sample types, outlined in Table 1 below.
Comparison of Plasma and Serum in Proteomics
|
Characteristic |
Plasma (anticoagulated blood) |
Serum (clotted blood) |
|
Composition |
Contains all blood proteins, including clotting factors (e.g. fibrinogen). |
Lacks clotting factors (fibrinogen largely absent; some coagulation proteins greatly reduced). |
|
Preparation method |
Draw blood into anticoagulant tube (EDTA, heparin, citrate), mix immediately, centrifuge to separate plasma. Quick to prepare (no wait time). |
Draw blood into plain tube, allow clotting ~30 min, then centrifuge to separate serum. Slower preparation due to clotting time. |
|
Handling considerations |
Less pre-analytical variability: immediate processing means minimal time at room temperature. Controlled procedure yields consistent results. |
More handling variability: requires controlled clotting time and temperature. If not standardized, clotting can vary between samples, affecting protein content. |
|
Protein content |
Includes high-abundance clotting proteins (fibrinogen ~0.3% of plasma by weight) and others that might be lost in clotting. |
Depleted in fibrinogen and some coagulation proteins (e.g. factor VII, IX, X, etc. are greatly reduced). Some proteins may be lower due to adsorption to the clot. |
|
Unique features |
Fewer artifacts from clotting – platelets remain intact, so less platelet activation/release of proteins (e.g. low platelet-derived VEGF). |
During clotting, cells (platelets, WBCs) can release proteins into serum. E.g., platelet-derived VEGF can be 6-fold higher in serum than plasma from the same blood. |
|
Stability |
Generally stable if processed quickly; long-term storage (frozen plasma) is possible for years with little degradation (often reported up to ~10 years). |
Slightly shorter shelf-life reported; some proteins may degrade or change during longer storage. Proper processing (prompt separation, freezing) is needed to maintain stability for months. |
|
Usage in proteomics |
Recommended by experts and consortia for proteomic studies (e.g. HUPO Plasma Proteome Project suggests EDTA plasma for proteomics). Common in plasma proteomics and biomarker studies. |
Historically used in clinical chemistry (many diagnostic blood tests use serum). Also used in proteomics, but awareness is growing that serum proteome may be altered by clotting process. |
Impact of Clotting on the Blood Proteome
Clotting doesn’t just remove proteins – it can also add or modify components in your sample. When blood clots, platelets and white blood cells become activated and release various factors into the surrounding fluid. This means certain proteins can show up at higher levels in serum than in plasma, purely due to the clotting process. A notable example is vascular endothelial growth factor (VEGF), a protein released by platelets: some studies found VEGF concentrations in normal individuals averaged 230 ± 63 pg/mL in serum but only 38 ± 8 pg/mL in plasma from the same donors (a ~6-fold increase in serum). This dramatic difference illustrates how clotting-induced platelet degranulation can skew the proteomic profile of serum.
Other differences include platelet factor 4 (a coagulation-related protein), which is lower in serum than plasma. In general, many coagulation factors (Factor VII, IX, X, XI, etc.) are either absent or significantly reduced in serum because they get consumed or trapped in the fibrin clot. Some of these factors might still be detectable at trace levels in serum, but plasma gives a more complete picture of them. On the flip side, some cellular proteins or inflammatory mediators might appear more in serum due to cell breakdown during clotting.
From a proteomics standpoint, these differences mean that serum and plasma are not interchangeable. If one is not careful, comparing proteomic data from serum vs. plasma could lead to misinterpretation. For instance, imagine a biomarker study comparing patient groups: if one study used serum and another used plasma, a protein like VEGF could seem “elevated” in one group simply because of sample type differences rather than true biology. Even within a study using serum, inconsistent clotting (time or temperature before centrifugation) might cause artifactual differences between samples. Researchers have cautioned that ex vivo clotting processes can introduce variability and false positives in differential expression analysis. In fact, inconsistencies in clot formation between samples can lead to false differential signals in serum proteomics (e.g. one sample clotted more completely and released more platelet proteins than another).
It’s important to note that some differences caused by clotting could overlap with real biology. For example, if a disease state causes patients’ blood to clot more rapidly or platelets to be hyperactive, those patients’ serum might consistently show higher levels of certain platelet-released proteins. In such a case, the serum proteomic difference is real and related to disease. However, to confidently link it to biology, one must ensure the sample handling is standardized across compared groups. Overall, the proteomics community increasingly recognizes that plasma and serum are distinct matrices – a nuance that was sometimes underestimated in earlier studies where the terms were even used interchangeably. Modern proteomic studies emphasize understanding these differences when designing experiments and interpreting data.
Why Plasma Proteomics Is Often Preferred
Given the above, many experts recommend plasma as the sample of choice for blood proteomics, especially in discovery research. Here are the key reasons why plasma proteomics is often favored (and why our conclusion leans toward plasma):
More Complete Protein Representation
Plasma retains the full complement of blood proteins, including clotting factors. Important biomarkers could reside in those coagulation proteins or be bound to them. Using plasma ensures you’re not unknowingly discarding that part of the proteome. The Human Proteome Organization’s Plasma Proteome Project (HPPP) recommended as far back as 2005 that EDTA plasma be the preferred sample type for all proteomic experiments. This recommendation was based on the recognition that plasma gives a more comprehensive view of the blood proteome. Today, indeed >70% of plasma/serum proteomics datasets in large projects use plasma.
Controlled Sampling and Less Degradation
Plasma is collected with an anticoagulant and processed immediately, which can minimize protein degradation or modification. There is no 30-minute wait at room temperature as with serum. This is crucial because proteases and other enzymes can become active in that interim period during serum preparation. Recent studies highlight that time and temperature have strong influence on the plasma/serum proteome – keeping blood even a couple of hours at 4°C or room temp can alter many proteins (e.g., by degradation or release from cells). By using plasma and rapidly spinning and freezing samples, you reduce the risk of such pre-analytical changes. In other words, plasma offers more consistent and stable handling: draw, mix, spin, and freeze can all happen in minutes.
Lower Variability Between Samples
Because serum preparation involves an inherent delay and a biological process (clotting) that can vary slightly between individuals and conditions, serum samples may introduce extra variability. Plasma preparation is more mechanical and easier to standardize. One publication noted that partial ex-vivo degradation and modification of proteins during serum processing is likely a reason serum can be less informative. By avoiding that degradation (with plasma), you get more reliable data. This can be especially important in clinical proteomics where consistent results are key.
Better Biomarker Discovery Performance
Perhaps the most compelling evidence comes from direct comparisons of proteomic performance. A recent study in AJOG Global Reports (2023) compared the predictive power of proteomic signatures derived from paired serum vs. plasma samples in pregnant women. The plasma proteomic signature outperformed the serum signature for predicting a clinical outcome (gestational age) – with higher predictive correlation (R=0.64 for plasma vs R=0.45 for serum) and stronger statistical significance. Moreover, when they tried to apply a serum-derived protein signature to plasma data, it worked well, but a plasma-derived signature applied to serum data lost some power. The authors concluded that “serum proteomics are less informative than plasma proteomics” and that plasma is the preferred matrix for such studies. In essence, important signals were easier to detect in plasma, likely because the serum processing had partially degraded or altered some proteins (supporting the idea that the clotting process diminishes data quality).
Consistency with Other Studies
Using plasma also aligns with the majority of large proteomic initiatives and biobanks, meaning your results will be more directly comparable with other data. For example, the UK Biobank Pharma Proteomics Project collected plasma on tens of thousands of participants for proteomic analysis, not serum. If you foresee comparing or integrating data from different sources, plasma might provide a common ground.
That said, serum is not without merit. Serum remains the standard for many routine clinical assays (such as metabolic panels, hormone tests, etc.) due to historical reasons and because many assays were developed on serum. From a proteomics perspective, serum can certainly be used – and indeed many proteomic studies have successfully used serum for biomarker discovery. A quick PubMed search shows thousands of proteomics papers on both “serum proteomics” and “plasma proteomics”. In fact, as of late 2021 the counts were almost equal (around 10,000+ papers each). This underscores that both sample types are workable if handled properly. However, the trend in the last 5 years has been a shift toward plasma for the reasons outlined. Plasma’s advantages in preserving the native protein state and reducing pre-analytical artifacts give it an edge for unbiased discovery work.
Serum vs. Plasma in Blood Proteomics: What Comparative Studies Reveal
For those interested in head-to-head comparisons, several studies provide data on how serum and plasma differ in proteomic outputs:
Proteins Identified and Quantified: One systematic evaluation (Lan et al., 2018) compared EDTA-plasma, heparin-plasma, and serum for shotgun proteomics. It found that EDTA and heparin plasma were quite similar in terms of total proteins identified and quantification precision, while serum showed a largely overlapping proteome but with some differences in specific proteins. Importantly, the study recommended not to mix plasma and serum within the same proteomic study, and to use a single type of sample for consistency. In other words, you can get robust results with either matrix, but you need to account for their differences and stick to one matrix in any given experiment.
Normalization Between Serum and Plasma: A very recent advance by Shraim et al. (2025) developed a method to mathematically transform proteomic data from one matrix to the other. By measuring 1,463 proteins in matched plasma and serum samples using an Olink high-throughput assay, they derived protein-specific conversion factors to normalize serum measurements to plasma-equivalent values (and vice-versa). They successfully generated conversion models for 686 proteins, and validated over 550 of these models in independent datasets. This work confirms that while serum and plasma measurements are correlated, many proteins have systematic differences in level between the two – hence the need for a conversion factor for each protein. The takeaway: if one must integrate serum and plasma data, such normalization is possible, but it’s far easier to just use one sample type. The authors noted that aligning serum with plasma data helps “enhance collaborative analyses” across studies. Implicitly, this also reinforces that plasma is seen as the reference standard (since they often converted serum values to plasma scale).
Extracellular Vesicle Proteomics: Even in niche areas like exosome or microvesicle analysis, the choice of serum vs plasma matters. For example, a study on small extracellular vesicles (sEVs) in mouse blood found that serum EV fractions contained more platelet-related proteins than plasma EV fractions (consistent with platelet activation during clotting). This highlights that any downstream proteomic component of blood can be affected by the initial sample type.
In summary, the data from recent research overwhelmingly support the notion that plasma provides a more stable and complete snapshot of the blood proteome, which can translate to better biomarker discovery and reproducibility. Serum can certainly be used, especially if plasma collection is not feasible for some reason, but one must be diligent in controlling pre-analytical variables.
Best Practices for Sample Handling in Blood Proteomics
If you are planning a blood proteomics study or biomarker project, here are some practical tips based on the latest insights:
1. Stick to One Sample Type: Consistency is critical. Whether you choose plasma or serum, use the same matrix across all samples in your study. Mixing sample types introduces confounding effects and compromises data comparability. Most experts recommend plasma, but if serum is used, apply it uniformly with consistent processing.
2. Standardize Collection Protocols: For plasma, choose one anticoagulant (e.g., EDTA, heparin, or citrate) and apply identical processing times. EDTA is commonly used in mass spectrometry-based proteomics because it chelates calcium to prevent coagulation and inhibits some proteases. Heparin and citrate are also acceptable, though heparin can sometimes interfere with downstream MS workflows.
3. Control Clotting for Serum: If you must use serum, ensure standardized clotting time (typically 30 minutes at room temperature), followed by immediate centrifugation and freezing. Variations in clotting time or temperature can alter protein profiles, especially due to platelet activation.
4. Minimize Processing Delays: Regardless of sample type, process blood promptly. Delays longer than 1 hour—even on ice—can lead to protein degradation or release from cells. Plasma offers an advantage here, as it can be processed immediately after collection. In contrast, serum requires a waiting period during clotting, which must be managed carefully.
5. Future-Proofing Your Study: If your proteomics project might scale to clinical trials or multi-center collaborations, plasma is generally easier to standardize across sites. It’s also more commonly used in multi-omics initiatives and large biobank studies, enhancing cross-study compatibility.
Reference:
1. Lan J, Núñez Galindo A, Doecke J, et al. Systematic Evaluation of the Use of Human Plasma and Serum for Mass-Spectrometry-Based Shotgun Proteomics. J Proteome Res. 2018;17(4):1426-1435. doi:10.1021/acs.jproteome.7b00788
2. Ignjatovic V, Geyer PE, Palaniappan KK, et al. Mass Spectrometry-Based Plasma Proteomics: Considerations from Sample Collection to Achieving Translational Data. J Proteome Res. 2019;18(12):4085-4097. doi:10.1021/acs.jproteome.9b00503
3. Zhang X, Takeuchi T, Takeda A, Mochizuki H, Nagai Y. Comparison of serum and plasma as a source of blood extracellular vesicles: Increased levels of platelet-derived particles in serum extracellular vesicle fractions alter content profiles from plasma extracellular vesicle fractions. PLoS One. 2022;17(6):e0270634. Published 2022 Jun 24. doi:10.1371/journal.pone.0270634
4. Gegner HM, Naake T, Dugourd A, et al. Pre-analytical processing of plasma and serum samples for combined proteome and metabolome analysis. Front Mol Biosci. 2022;9:961448. Published 2022 Dec 20. doi:10.3389/fmolb.2022.961448
5. Espinosa C, Ali SM, Khan W, et al. Comparative predictive power of serum vs plasma proteomic signatures in feto-maternal medicine. AJOG Glob Rep. 2023;3(3):100244. Published 2023 Jun 12. doi:10.1016/j.xagr.2023.100244
6. 3. Shraim R, Diorio C, Canna SW, et al. A Method for Comparing Proteins Measured in Serum and Plasma by Olink Proximity Extension Assay. Mol Cell Proteomics. Published online May 27, 2025. doi:10.1016/j.mcpro.2025.101000
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


