Glycoproteomics: Mass Spectrometry-Based Insights into Protein Glycosylation in Health and Disease
Protein glycosylation is one of the most abundant and functionally important post-translational modifications (PTMs) in biology. Glycoproteins regulate cell–cell communication, immune recognition, receptor signaling, metabolism, and host–pathogen interactions. Aberrant glycosylation patterns are closely associated with cancer, cardiovascular disease, neurological disorders, kidney disease, metabolic disease, and viral infection.
In this overview, we introduce the basic concepts of glycoproteomics, summarize LC–MS/MS-based glycoproteomics workflows, and highlight how site-specific glycoprotein biomarkers are being used in clinical and translational research.
Glycoproteomics is the branch of proteomics that systematically studies glycosylated proteins on a proteome-wide scale. Using advanced liquid chromatography–tandem mass spectrometry (LC–MS/MS) and bioinformatics, glycoproteomics aims to:
- Identify glycoproteins and glycosylation sites
- Characterize N-glycans and O-glycans and their structural heterogeneity
- Quantify site-specific glycosylation changes across biological conditions
This site-specific, mass-spectrometry-based clinical glycoproteomics is emerging as a powerful strategy for biomarker discovery and precision medicine.
What Is Glycoproteomics?
Glycoproteomics focuses on intact glycopeptides: peptides that still carry their attached glycans. By mapping peptide sequence, glycosylation site, and glycan composition in a single LC–MS/MS experiment, researchers can capture the full complexity of the glycoproteome.
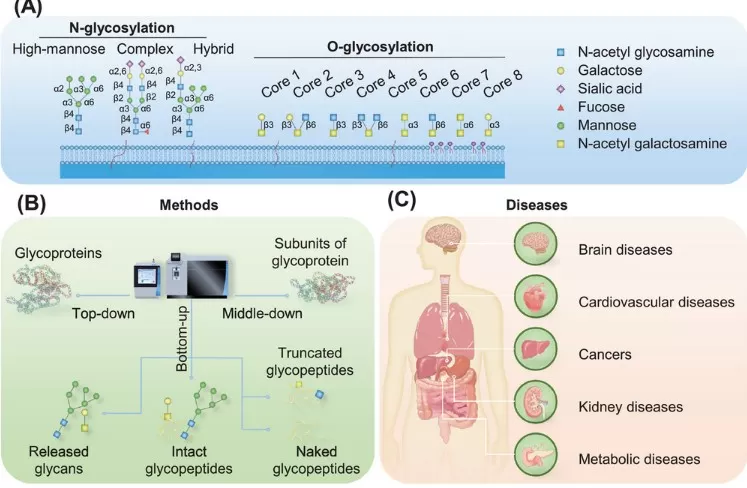
Figure 1. Overview of Protein Glycosylation and Intact Glycopeptides
Two key concepts are:
Macroheterogeneity – whether a particular site is glycosylated or not (site occupancy)
Microheterogeneity – which glycan structures occupy a given glycosylation site and in what proportions
This microheterogeneity reflects variations in monosaccharide composition, branching, linkage type, and terminal modifications (e.g., fucosylation, sialylation), and is often disease-specific.
Modern mass-spectrometry-based glycoproteomics leverages:
- High-resolution instruments (e.g., Orbitrap, TOF)
- Optimized fragmentation methods (HCD, stepped-HCD, ETD/EThcD)
- Dedicated search engines for intact glycopeptide analysis
Together, these technologies enable system-wide, site-specific N-glycoproteomics and O-glycoproteomics in complex clinical samples.
Major Types of Protein Glycosylation
The two dominant forms of protein glycosylation studied in glycoproteomics are N-linked glycosylation and O-linked glycosylation.
|
Glycosylation type |
Attachment site |
Typical structures / subclasses |
Key functions & disease links |
|
N-glycosylation |
Asn in N-X-S/T motif (X ≠ Pro) |
High-mannose, complex, hybrid N-glycans |
Protein folding, stability, trafficking; altered in cancer, CVD, CKD, diabetes |
|
O-glycosylation |
Ser/Thr (no strict consensus sequence) |
Eight core O-glycan structures (e.g., core 1/2 O-GalNAc, sialyl-T/Tn, etc.) |
Mucin biology, immune regulation, lipoprotein metabolism, viral infection |
Changes in N-glycan branching, core fucosylation, and sialylation, or in O-glycan extension and truncation, are frequently observed in tumors, cardiovascular disease, autoimmune kidney diseases, and metabolic disorders.
Glycoproteomics Strategies: Top-Down, Middle-Down, Bottom-Up
Depending on how proteins are processed prior to MS analysis, glycoproteomics strategies can be grouped into:
- Top-down glycoproteomics – analyzes intact glycoproteins; maximizes structural context but is technically challenging and low throughput
- Middle-down glycoproteomics – studies large subunits or protein domains after limited proteolysis
- Bottom-up glycoproteomics (most common) – digests proteins into peptides and intact glycopeptides (IGPs) for LC–MS/MS
Bottom-up, LC–MS/MS-based intact glycopeptide profiling currently offers the best balance between depth of coverage, throughput, and applicability to complex clinical cohorts.
LC–MS/MS-Based Clinical Glycoproteomics Workflow
A typical LC–MS/MS-based clinical glycoproteomics workflow comprises five key stages, from study design and sample preparation to data analysis and biomarker validation.
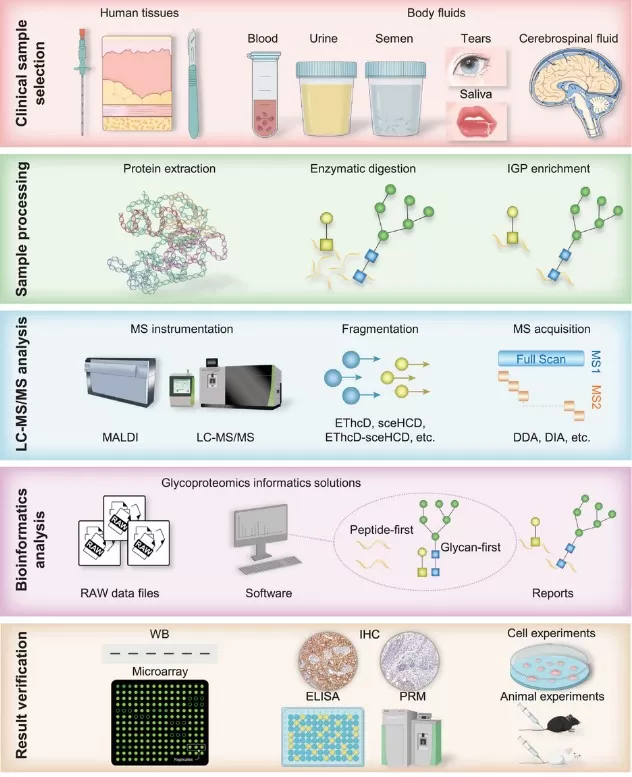
Figure 2. LC–MS/MS Workflow for Clinical Glycoproteomics
1. Clinical sample selection and study design
2. Sample preparation and intact glycopeptide enrichment
4. Bioinformatics and glycoproteomics data analysis
5. Experimental validation of glycoprotein biomarkers
1. Clinical Sample Selection and Study Design
For robust glycoprotein biomarker discovery, careful study design is essential:
1) Define disease subtype and stage (e.g., early vs late cancer, HF vs control)
2) Select appropriate sample type:
- Tissue biopsies (tumor vs adjacent normal)
- Biofluids: plasma/serum, urine, CSF, saliva, tears, semen
3) Ensure adequate cohort size and well-matched controls
4) Standardize collection, pre-processing, storage, and transport
Standard operating procedures help reduce pre-analytical variation and improve reproducibility in clinical LC–MS/MS glycoproteomics.
2. Sample Preparation and Intact Glycopeptide Enrichment
Sample preparation typically includes:
1) Protein extraction and solubilization
- Homogenization of tissue or biofluid
- Minimization of protein degradation and contamination
- For plasma/serum, depletion of high-abundance proteins to enhance detection of low-abundance glycoproteins
2) Enzymatic digestion
- Trypsin is the most widely used protease in bottom-up glycoproteomics
- Alternative or complementary proteases can be used to improve coverage of glycosylation sites
3) Intact glycopeptide (IGP) enrichment
Because glycopeptides are usually low in abundance, enrichment is critical, common strategies include:
- Hydrophilic interaction chromatography (HILIC)
- Lectin affinity chromatography (e.g., ConA, WGA, AAL)
- Mixed-mode and multi-step enrichment for broader coverage
The choice of enrichment material affects selectivity and bias and must be optimized for each sample type and study objective.
3. LC–MS/MS Analysis
Enriched intact glycopeptides are separated by nano-LC and analyzed on high-resolution mass spectrometers (e.g., Orbitrap, Q-TOF). Key considerations include:
- Ionization settings (commonly ESI) and gradient design
- MS/MS fragmentation modes: HCD, stepped HCD, ETD/EThcD, or hybrid methods
- Data-dependent acquisition (DDA) vs data-independent acquisition (DIA) for deeper and more reproducible coverage
Recent advances in LC–MS/MS instrumentation and acquisition strategies are pushing site-specific glycoproteomics closer to routine clinical use.
4. Bioinformatics and Glycoproteomics Data Analysis
Glycoproteomics data analysis is computationally demanding because each MS/MS spectrum must be explained by both a peptide backbone and a glycan structure. Current software can be broadly grouped into:
- Peptide-first search engines: Identify peptide sequences first, then assign glycans
- Glycan-first or hybrid search tools: Use glycan fragment ions as anchors in the search
These tools support:
- Identification of intact N- and O-glycopeptides
- Assignment of glycosylation sites
- Relative or absolute quantitative glycoproteomics across large cohorts
Standardized search parameters, scoring schemes, and reporting formats are being developed to improve reproducibility and comparability between laboratories.
5. Experimental Validation
Finally, candidate glycoprotein biomarkers and altered glycosylation sites must be validated by:
- Targeted MS assays (MRM/PRM for glycopeptides)
- Lectin blotting, ELISA, or antibody-based assays
- Functional studies in cell lines, organoids, and animal models
Multi-center, retrospective and prospective clinical studies are needed before glycoproteomics signatures can be translated into routine diagnostic tests.
Key Challenges in Glycoproteomics
Despite major progress, clinical glycoproteomics still faces significant technical and translational challenges.
1. Structural Complexity and Glycan Heterogeneity
- N- and O-glycans exhibit extreme structural heterogeneity (branching, linkages, isomers)
- O-glycosylation often lacks consensus motifs and universal glycosidases, making it harder to map than N-glycosylation
- Low abundance and microheterogeneity of glycopeptides complicate quantification in complex clinical samples
Emerging strategies include:
- Improved fragmentation (e.g., EThcD, stepped HCD) for richer glycan and peptide fragment coverage
- Orthogonal separation and acquisition schemes to decouple peptide and glycan information
- Better glycopeptide enrichment tailored to different sample types
2. Data Analysis and Software Bottlenecks
Challenges in glycoproteomics data analysis include:
- Ambiguous assignment of glycans to specific sites
- Limited reproducibility in quantitative glycoproteomics
- High computational cost for large-scale clinical studies
Ongoing efforts focus on:
- New algorithms for modular interpretation of peptide and glycan fragments
- Standardized, community-approved data-processing pipelines and benchmarking datasets
- Cloud-based and high-performance computing for high-throughput glycoproteomics analysis
3. Translational Barriers and Standardization
For true clinical implementation, glycoproteomics methods must be:
- Robust and scalable for routine diagnostics laboratories
- Compatible with regulatory requirements and external quality assessment (EQA) schemes
- Supported by clear guidelines for sample handling, data analysis, and result reporting
Standardization of clinical glycoproteomics workflows will be key to translating research findings into validated glycoprotein biomarkers.
Clinical Applications of Glycoproteomics in Disease Research
Altered glycoproteomes are now recognized as hallmarks of many major human diseases. Mass-spectrometry-based clinical glycoproteomics enables the discovery of disease-specific glycoprotein signatures and glycosylation-based biomarkers, offering new tools for diagnosis, prognosis, and patient stratification.
Neurological Disorders and Alzheimer’s Disease
In neurological disorders, glycoproteomics has revealed how abnormal protein glycosylation contributes to neurodegeneration and psychiatric disease. In Alzheimer’s disease (AD), the microtubule-associated protein tau is a critical glycoprotein whose aberrant glycosylation in the Golgi apparatus promotes tau accumulation, amyloid-β plaque formation, and neurofibrillary tangle development, ultimately driving neuronal loss and cognitive decline. Beyond AD, glycoproteomics studies have identified altered N-glycosylation patterns in proteins associated with Parkinson’s disease and have shown that abnormal N-glycosylation of GABA receptor subunits in schizophrenia may influence synaptic transmission and neuropsychiatric phenotypes. Together, these findings highlight the potential of brain glycoproteomics and cerebrospinal fluid glycopeptide profiling for uncovering biomarkers and mechanisms in neurodegenerative and psychiatric disorders.
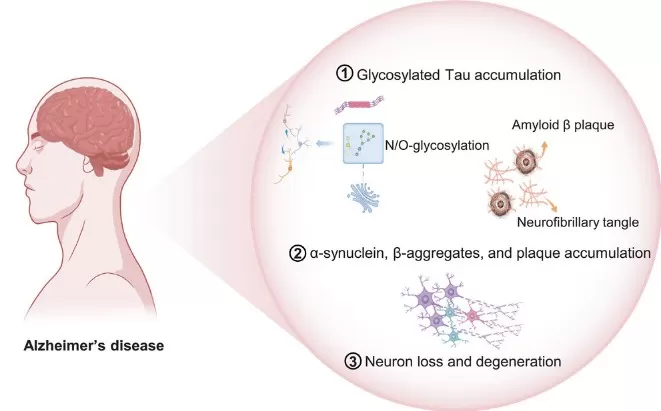
Figure 3. Pathogenesis of Alzheimer’s Disease
Key glycoproteomics insights in neurology:
- Tau glycosylation promotes aggregation and pathology in Alzheimer’s disease.
- N-glycosylation changes are observed in Parkinson’s disease–related proteins.
- Abnormal N-glycosylation of GABA receptors is linked to schizophrenia.
Cardiovascular Disease (CVD)
In cardiovascular disease, glycosylation modulates lipid metabolism, inflammation, thrombosis, and cardiac remodeling, making glycoproteins attractive candidates for mechanistic studies and biomarker discovery. In atherosclerosis, O-glycosylation of PSGL-1 by GALNT4 enhances monocyte adhesion to the endothelium and accelerates plaque development, while platelet glycoprotein VI (GPVI) and glycated lipoproteins have emerged as potential biomarkers of thrombotic risk. In heart failure (HF), N-glycosylation of the transmembrane glycoprotein CD147 influences cardiac hypertrophy and fibrosis, whereas core fucosylation of β1-adrenergic receptors (β1AR) modulates cAMP/PKA signaling and myocardial contractility. Ischemic heart disease (IHD) is also associated with dynamic changes in the glycosylation of apolipoproteins such as ApoJ during ischemia and reperfusion. These examples illustrate how cardiovascular glycoproteomics can identify glycan-dependent mechanisms and support the development of glycoprotein-based biomarkers and glycosylation enzymes as therapeutic targets.
Key glycoproteomics insights in CVD:
- PSGL-1 O-glycosylation promotes monocyte adhesion and atherogenesis.
- CD147 and β1AR N-glycosylation regulate cardiac remodeling and function.
- Apolipoprotein glycosylation changes during ischemia–reperfusion in IHD.
Cancer Glycoproteomics
Cancer glycoproteomics dissects how tumor cells remodel their glycome and glycoproteome to support malignant growth, invasion, immune evasion, and therapy resistance. In lung cancer (NSCLC), specific N-glycosylation and fucosylation patterns on EGFR correlate with resistance to tyrosine kinase inhibitors, while salivary and exosomal glycoproteins such as lactoferrin, MUC5B, and CD63 have been proposed as non-invasive biomarkers. In colorectal cancer (CRC), altered glycosylation of adhesion and metabolism-related proteins, including ICAM1 and APMAP, has been linked to metastasis, and dysregulated expression of O-glycosyltransferases such as C1GALT1 and B4GALNT2 is associated with clinical outcome. In hepatocellular carcinoma (HCC), enhanced core fucosylation of serum IgG, IgA1, and the folate receptor FOLR1 in HBV-related disease, as well as urinary exosomal N-glycopeptides, have been reported as markers of metastasis and poor prognosis. For pancreatic ductal adenocarcinoma (PDAC), the classic glycan marker CA19-9 (sialyl-Lewis a) lacks specificity, but combining it with fucosylated SERPINA1 and IgG galactosylation patterns improves detection, especially in CA19-9-negative patients. Collectively, these studies demonstrate how site-specific cancer glycoproteomics can refine tumor classification, stratify patients, and guide targeted therapy.
Key glycoproteomics insights in cancer:
- EGFR N-glycosylation and fucosylation are linked to drug resistance in NSCLC.
- ICAM1/APMAP glycosylation and O-glycosyltransferase expression inform CRC metastasis and prognosis.
- Core-fucosylated immunoglobulins and exosomal N-glycopeptides serve as biomarkers in HCC and PDAC.
Kidney Diseases
Glycoproteomics has transformed our understanding of several major kidney diseases by revealing how aberrant glycosylation affects immune complexes, autoantibodies, and tubular injury. In IgA nephropathy (IgAN), galactose-deficient IgA1 (Gd-IgA1) with aberrant O-glycosylation in the hinge region is central to disease pathogenesis, and lectin-based assays combined with glycopeptide profiling are now used to quantify Gd-IgA1 as a non-invasive biomarker. In diabetic nephropathy (DN), stage-specific N-glycoprotein signatures in urine and plasma have been identified, and up-regulation of zinc-α2-glycoprotein (ZAG) appears as an early indicator of renal damage even before macro-albuminuria develops. In membranous nephropathy (MN), abnormal glycosylation of IgG4 autoantibodies against PLA2R1—including changes in sialylation, core fucosylation, and galactosylation—distinguishes different disease states, and combinations of specific N-glycan structures enable molecular subtyping of PLA2R1-related MN. These examples underscore the value of site-specific quantitative N-glycoproteomics for the diagnosis, prognosis, and molecular classification of kidney diseases.
Key glycoproteomics insights in kidney disease:
- Gd-IgA1 O-glycosylation is a central biomarker and driver of IgA nephropathy.
- Urinary and plasma N-glycoprotein signatures, including ZAG, mark early diabetic nephropathy.
- IgG4 glycosylation patterns refine molecular subtyping of PLA2R1-related membranous nephropathy.
Diabetes, Obesity, and Metabolic Disease
In diabetes and obesity, chronic low-grade inflammation and metabolic dysregulation are tightly linked to changes in the IgG glycome and the circulating glycoproteome. In type 1 diabetes (T1D), increased high-mannose and bisecting GlcNAc N-glycans together with decreased IgG galactosylation form characteristic glycan patterns that can help predict disease risk. In type 2 diabetes (T2D), IgG galactosylation levels correlate with the presence and severity of diabetic nephropathy, retinopathy, and cardiovascular events, while complement proteins such as C1s and C4 exhibit altered N-glycosylation. Obesity further shifts the IgG glycome toward a more pro-inflammatory profile. Importantly, lifestyle and interventional studies have shown that dietary restriction, bariatric surgery, and structured exercise can remodel N-glycan branching, galactosylation, and sialylation in directions consistent with a “younger” GlycanAge, suggesting that glycosylation patterns may be both biomarkers and mediators of metabolic health.
Key glycoproteomics insights in metabolic disease:
- IgG N-glycan patterns predict and track complications in T1D and T2D.
- Complement protein glycosylation is altered in diabetic complications.
- Weight loss and exercise can rejuvenate the IgG glycome and lower GlycanAge.
Viral Infection and COVID-19
Viral infections, especially COVID-19, have highlighted the importance of viral and host glycoproteomics for understanding pathogenesis and immune responses. The SARS-CoV-2 spike protein carries numerous N- and O-glycans that facilitate binding to the ACE2 receptor, shield key epitopes from neutralizing antibodies, and modulate tissue tropism and immune escape. Glycoproteomics has been used to map spike glycosylation sites in detail and to monitor how viral variants or vaccines alter glycan occupancy and processing. At the same time, profiling of host immunoglobulin glycosylation—such as reduced fucosylation of anti–SARS-CoV-2 IgG—has revealed associations with disease severity and hyperinflammatory responses. These findings support the development of glyco-informed vaccines, therapeutic antibodies, and antiviral strategies that account for both viral and host glycosylation.
Key glycoproteomics insights in viral infection:
- Spike protein N- and O-glycans regulate ACE2 binding, immune evasion, and tropism.
- IgG glycosylation patterns correlate with COVID-19 severity and inflammatory responses.
- Viral and host glycoproteomics guide the design of vaccines and therapeutic antibodies.
Frequently Asked Questions about Glycoproteomics
Q1. What is glycoproteomics used for?
Glycoproteomics is primarily used to map site-specific N- and O-glycosylation, discover disease-specific glycoprotein biomarkers, and understand how glycosylation affects protein function in health and disease.
Q2. What techniques are commonly used in glycoproteomics?
Most glycoproteomics workflows rely on LC–MS/MS analysis of enriched intact glycopeptides, combined with high-resolution mass spectrometry and dedicated software tools for peptide–glycan identification and quantification.
Q3. Which diseases benefit most from glycoproteomics studies?
Glycoproteomics is being actively applied to cancer, cardiovascular disease, kidney disease, neurodegenerative disorders, diabetes and obesity, as well as infectious diseases such as COVID-19.
Take-Home Messages
- Glycoproteomics systematically characterizes protein glycosylation at the proteome level, with a strong focus on site-specific N- and O-glycosylation.
- Modern LC–MS/MS-based glycoproteomics workflows that combine intact glycopeptide enrichment, high-resolution MS, and dedicated bioinformatics tools enable deep and quantitative profiling in clinical samples.
- Disease-specific changes in glycosylation across neurological disorders, cardiovascular disease, cancer, kidney disease, diabetes, obesity, and infectious diseases make glycoprotein biomarkers highly attractive for diagnosis, prognosis, and patient stratification.
- Despite challenges in structural heterogeneity, data analysis, and clinical standardization, rapid advances are bringing clinical glycoproteomics closer to real-world applications in precision medicine.
Future Directions: From Glycoproteomics to Precision Medicine
Looking ahead, several trends are shaping the future of clinical glycoproteomics:
Multi-omics integration
Combining glycoproteomics with proteomics, glycomics, transcriptomics, metabolomics, and genomics to model glycosylation networks and regulatory pathways.
AI and machine learning
Using computational models to predict glycosylation sites, infer structure–function relationships, and discover glycoprotein signatures for patient stratification.
Standardized, quantitative pipelines
Developing robust, site-specific quantitative N- and O-glycoproteomics workflows that are ready for large clinical cohorts and longitudinal studies.
Biopharmaceutical applications
Applying glyco-engineering to therapeutic antibodies, fusion proteins, and vaccines to optimize efficacy, stability, and safety based on defined glycan profiles.
As instrumentation, software, and clinical validation mature, mass-spectrometry-based glycoproteomics is poised to become a routine component of precision medicine. For groups interested in applying these approaches, MetwareBio’s N-glycosylation proteomics service offers site-specific LC–MS/MS workflows and bioinformatics support to profile glycoprotein biomarkers in clinical and translational research.
Reference
Wang Y, Lei K, Zhao L, Zhang Y. Clinical glycoproteomics: methods and diseases. MedComm (2020). 2024;5(10):e760. Published 2024 Oct 4. doi:10.1002/mco2.760
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


