DIP-MS: Ultra-Deep Proteomics Unravels Protein Complexes
Protein complexes are macromolecular assemblies essential for cellular processes, where multiple proteins interact to form functional units that regulate biochemical pathways, signal transduction, and other key cellular functions. These complexes are ubiquitous in organisms, acting as dynamic structures that catalyze and coordinate biological activities. For instance, they play pivotal roles in disease mechanisms, such as in neuropsychiatric disorders where proteins interact in pathways affecting neurological functions. Studying protein complexes is critical because their dysfunction underlies a wide range of human diseases, including metabolic disorders, psychiatric conditions, and infectious diseases like COVID-19. By understanding how complexes assemble and function, researchers can unravel complex phenotypes such as obesity, where specific protein assemblies on extracellular vesicles influence adiposity traits and correlate with body fat levels. This knowledge not only elucidates disease etiology but also opens avenues for targeted therapies.
Research on protein complexes yields significant clinical and therapeutic implications. By mapping protein quantitative trait loci (pQTL), studies capture up to 50% of each protein's heritability, revealing causal links to diseases like hypertension, high cholesterol, and psychiatric disorders. For instance, proteogenomic resource efforts have validated drug-target combinations—such as identifying existing drugs for repurposing and highlighting new therapeutic avenues—providing a foundation for precision medicine. Additionally, genetic insights into complexes on extracellular vesicles demonstrate their role in obesity and lipid metabolism, with EVs carrying specific surface proteins correlating negatively with body adiposity and enhancing heritability enrichment for cholesterol levels. This guides biomarker development for monitoring disease progression. Overall, studying protein complexes not only advances understanding of modular proteome architecture but also drives innovations in diagnosis and treatment.
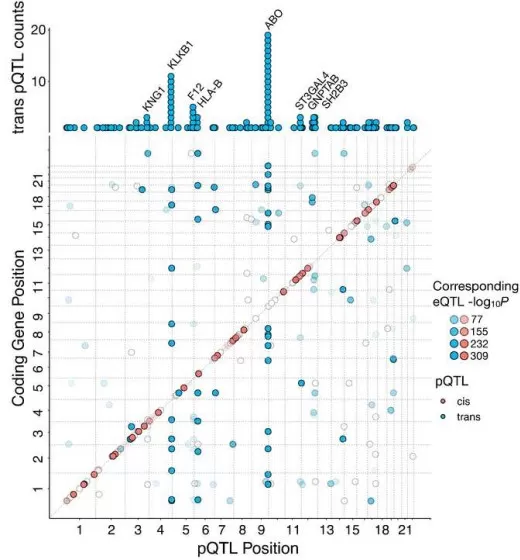
overlap of the pleiotropic mapped pQTL with existing eQTL
Protein complexes drive most cellular processes, yet resolving their composition and dynamics has long been a challenge. To analyze protein complexes, advanced methods leverage mass spectrometry and genetic techniques. Affinity Purification combined with Mass Spectrometry (AP-MS) has been a cornerstone, where proteins of interest are purified and their interacting partners identified via tandem mass spectrometry. However, traditional AP-MS struggles to resolve complex isoforms in a single experiment, often requiring time-consuming reciprocal approaches for reliable inference. Innovative methods like Deep Interactome Profiling by Mass Spectrometry (DIP-MS) overcome this by integrating multiple separation and acquisition techniques. A recent study published in ‘Nature Methods’ (2024) introduces DIP-MS (Deep Interactome Profiling by Mass Spectrometry), a novel method that enables high-resolution deconvolution of protein complexes in a single experiment. By integrating affinity purification, blue-native PAGE, data-independent acquisition (DIA-MS), and deep-learning analysis (PPIprophet), DIP-MS significantly improves the depth and accuracy of protein complex profiling. Applied to the human prefoldin family, this technique uncovered new complex isoforms and subunits, offering an unprecedented view into the modular organization of the proteome.
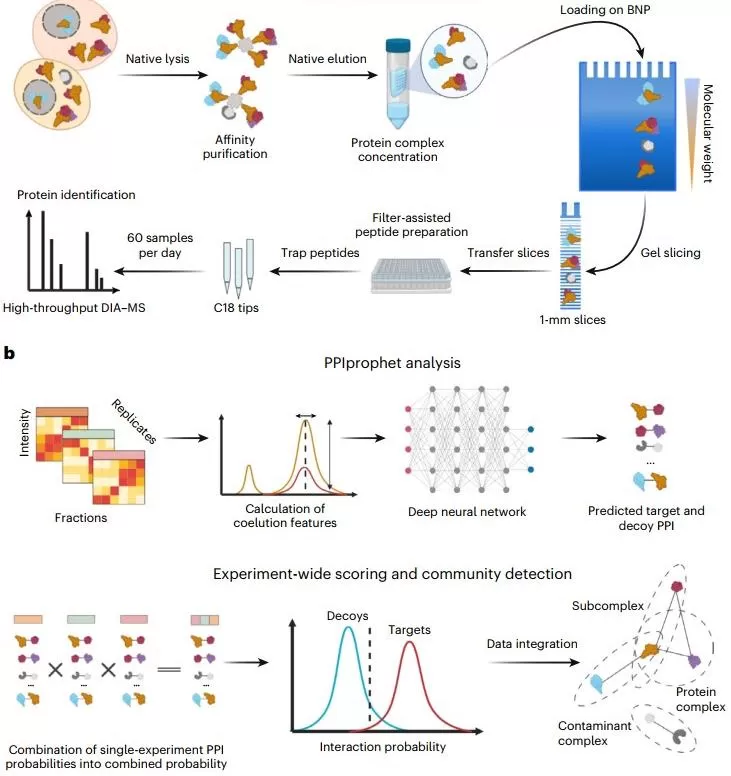
Experimental and computational DIP-MS workflow
One-shot resolution of multiple complex isoforms
DIP-MS separates bait-associated complexes via BN-PAGE and identifies coeluting proteins through DIA-MS. Using the deep-learning tool PPIprophet, the method predicts true interactions and complex architectures. This allows researchers to resolve multiple distinct complexes sharing a common bait, without requiring reciprocal AP–MS runs.
DIP-MS outperforms traditional interactomics workflows
Compared to SEC–MS and AP–MS, DIP-MS identified 187 more interactors using PFDN2 as bait, recovering 30% of known prefoldin-related interactions. It achieved higher recall while maintaining comparable precision and required significantly less sample input and time, highlighting its efficiency and sensitivity.
Discovery of novel prefoldin complexes
DIP-MS not only confirmed canonical prefoldin and prefoldin-like assemblies but also revealed a new PDRG1-containing prefoldin homolog complex. It identified POLR2E as a core subunit of the PFDL complex and mapped client interactions, subassemblies, and stoichiometry—discoveries that were validated by structure prediction and AP–MS.
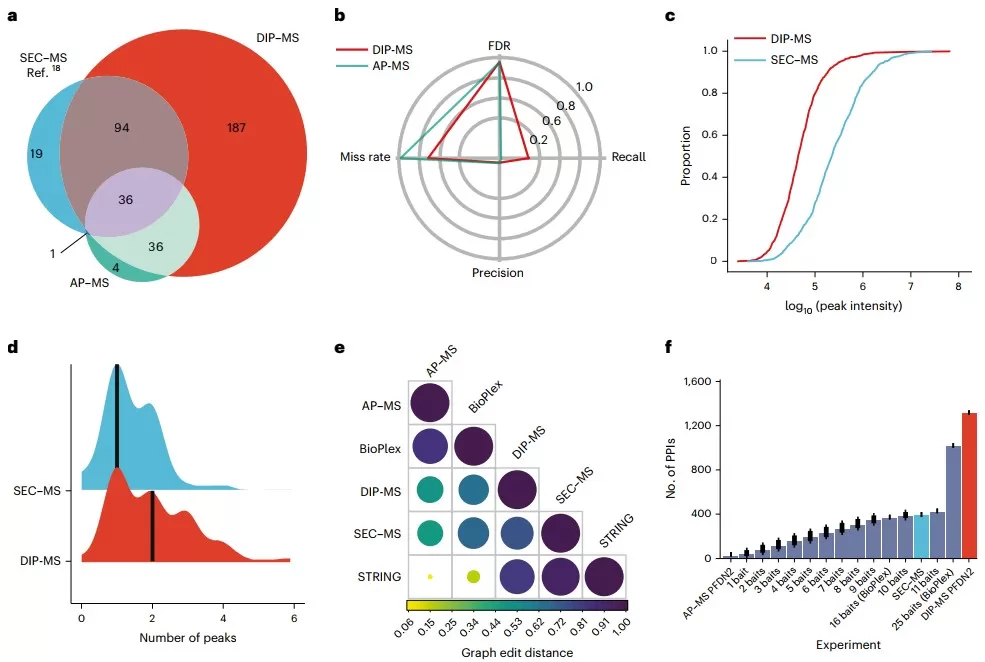
Benchmark of DIP-MS versus other techniques for interactome analysis
DIP-MS sets a new benchmark for proteomics by enabling single-experiment mapping of protein interaction networks with high resolution. It consolidates years of interactome studies and opens doors to dynamic, condition-specific complex analysis. The integration of native separation, DIA-MS, and machine learning makes DIP-MS ideal for studying proteostasis, signaling, and disease-associated assemblies. As part of multi-omics strategies, it provides foundational insight into proteome architecture.
Explore Proteomics with MetwareBio
MetwareBio offers comprehensive proteomics and multi-omics services, including DIA-based global profiling, PTM analysis, and complex-centric workflows. Powered by state-of-the-art mass spectrometry platforms and expert bioinformatics, our one-stop solutions help researchers achieve publication-ready insights. Contact us to learn more and accelerate your interactome discovery journey.
References:
- Frommelt F, et al., The genetic landscape of neuro-related proteins in human plasma. Nat Hum Behav. 2024 Nov;8(11):2222-2234.
- Repetto L, et al., Unraveling Neuro-Proteogenomic Landscape and Therapeutic Implications for Human Behaviors and Psychiatric Disorders. Nat Hum Behav. 2024 Nov;8(11):2222-2234. doi:10.1038/s41562-024-01963-z.
- Frommelt F, Fossati A, Uliana F, Wendt F, Xue P, Heusel M, Wollscheid B, Aebersold R, Ciuffa R, Gstaiger M. DIP-MS: ultra-deep interaction proteomics for the deconvolution of protein complexes. Nat Methods. 2024 Apr;21(4):635-647.
Read more
- Protein Complexes: What They Are and Why They Matter in Biomedical Research
- Understanding WGCNA Analysis in Publications
- Beginner for KEGG Pathway Analysis: The Complete Guide
- GSEA Enrichment Analysis: A Quick Guide to Understanding and Applying Gene Set Enrichment Analysis
- LC-MS VS GC-MS: What's the Difference
- LC vs. HPLC vs. UHPLC: Tracing the Evolution of Chromatographic Techniques
- Mastering Chromatography: Everything You Need to Know
- DIA Proteomics vs DDA Proteomics: A Comprehensive Comparison
- Malic Acid vs. Citric Acid: The Powerhouse Acids in Your Favorite Fruits
- Fumaric Acid Unveiled: From Nature's Palette to Therapeutic Potential
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


