What Is Discovery Proteomics? A Practical Guide to Workflow, Data Analysis, and Applications
Comprehensive analysis of protein expression is essential for understanding biological mechanisms, disease progression, and drug response. As proteomics studies continue to scale in sample number and complexity, researchers increasingly rely on LC–MS/MS–based discovery proteomics to achieve broad proteome coverage, quantitative consistency, and reproducible results across experiments.
Discovery proteomics enables large-scale protein profiling through standardized experimental workflows and advanced data analysis. By supporting unbiased protein identification, quantitative comparison, and pathway-level interpretation, it has become a widely adopted approach for exploratory research and early-stage discovery studies. In this guide, we provide a practical overview of the discovery proteomics workflow, data analysis considerations, and key application areas, offering a clear framework for researchers evaluating discovery proteomics strategies for their own studies.
What Is Discovery Proteomics?
Discovery proteomics is an unbiased, LC–MS/MS–based approach designed to answer a fundamental biological question: which proteins are present in a sample, and how do their abundances change across different biological conditions?
Often referred to as shotgun proteomics or global proteomics, discovery proteomics enables the simultaneous identification and quantification of thousands of proteins within a single experiment. By avoiding predefined target selection, this approach provides broad proteome coverage and supports comprehensive comparison of protein expression across samples.
As a result, discovery proteomics is widely applied to disease versus control studies, drug response and mechanism-of-action analysis, pathway and network exploration, and early-stage biomarker discovery. Its unbiased nature makes it particularly well suited for exploratory research and hypothesis generation, especially when the key molecular drivers of a biological system are not yet fully characterized.
Discovery Proteomics vs Targeted Proteomics
While discovery proteomics and targeted proteomics address different experimental goals, they are often used as complementary strategies within a single proteomics pipeline, supporting both exploratory discovery and hypothesis-driven validation.
Discovery proteomics is designed for unbiased, proteome-wide profiling, enabling the identification and quantification of thousands of proteins without predefined targets. This global coverage makes it particularly well suited for exploratory studies, such as comparative proteome analysis, pathway discovery, and early-stage biomarker identification, where the relevant proteins and biological drivers are not yet known.
In contrast, targeted proteomics approaches—such as selected reaction monitoring (SRM) and parallel reaction monitoring (PRM)—focus on a predefined set of peptides or proteins. By restricting data acquisition to specific targets, targeted proteomics achieves higher sensitivity, quantitative precision, and reproducibility, making it ideal for validation, verification, and routine quantification of known protein markers across large sample sets. (Learn more at: PRM vs MRM: A Comparative Guide to Targeted Quantification in Mass Spectrometry)
From a workflow perspective, discovery proteomics is typically positioned at the front end of proteomics research, where it generates comprehensive protein profiles and candidate lists. These candidates are then refined, prioritized, and quantitatively validated using targeted proteomics in follow-up studies. Together, discovery and targeted proteomics form an integrated strategy that balances breadth and depth, supporting robust biological discovery and translational research.
Comparison Between Discovery and Targeted Proteomics
|
Aspect |
Discovery Proteomics |
Targeted Proteomics |
|
Primary objective |
Global protein identification and relative quantification |
Precise quantification of predefined proteins |
|
Target selection |
Unbiased, no prior target definition required |
Requires predefined protein or peptide targets |
|
Proteome coverage |
Thousands of proteins per experiment |
Dozens to hundreds of proteins |
|
Common acquisition modes |
Data-dependent acquisition (DDA), data-independent acquisition (DIA, e.g., SWATH-MS) |
Selected/multiple reaction monitoring (SRM/MRM), parallel reaction monitoring (PRM) |
|
Sensitivity for low-abundance proteins |
Moderate |
High |
|
Quantitative precision |
Moderate to high (typically higher with DIA) |
Very high |
|
Missing values across runs |
Common in DDA, reduced in DIA |
Minimal |
|
Throughput and scalability |
Well suited for large-scale and cohort-based studies |
Best suited for focused validation studies |
|
Typical applications |
Exploratory research, pathway analysis, biomarker discovery |
Biomarker validation, targeted hypothesis testing, clinical assays |
Experimental Design Considerations in Discovery Proteomics
Careful experimental design is critical for generating reliable and biologically meaningful discovery proteomics data. Because discovery proteomics aims to capture global protein expression changes, variability introduced during sample collection, preparation, or acquisition can substantially affect downstream analysis and interpretation.
Key considerations include biological replication, sample randomization, and appropriate control selection. Sufficient biological replicates are essential to distinguish true biological variation from technical noise, particularly in studies involving complex tissues or heterogeneous clinical samples. Randomizing sample processing order and LC–MS/MS acquisition can help minimize batch effects, while the inclusion of quality control samples supports performance monitoring across large experiments.
Experimental design decisions also influence the choice of quantification strategy. Label-free approaches offer flexibility and scalability, whereas labeling-based methods such as TMT enable multiplexed analysis but require careful control of ratio compression and batch effects. Aligning experimental design with study objectives is therefore a foundational step in successful discovery proteomics workflows.
Discovery Proteomics Workflow
Discovery proteomics follows a standardized yet flexible workflow that integrates experimental design, mass spectrometry, and computational analysis. Each step plays a critical role in determining data quality, depth of coverage, and biological interpretability.
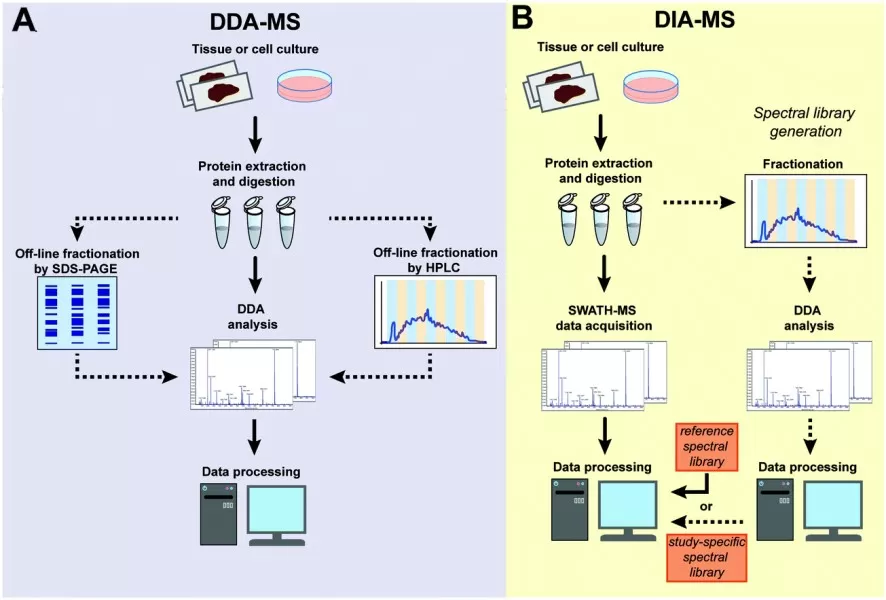
Overview of the discovery proteomics workflow using DDA and DIA acquisition strategies.
Image reproduced from Krasny and Huang, 2021, Molecular Omics, licensed under the Creative Commons Attribution 3.0 International License (CC BY 3.0).
Sample Preparation
The workflow begins with careful sample preparation, which directly influences proteome coverage and quantitative accuracy. Biological samples such as cells, tissues, or biofluids are first lysed to release proteins. The extracted proteins are then cleaned to remove interfering substances and enzymatically digested—most commonly using trypsin—into peptides that are suitable for mass spectrometry analysis.
Most discovery proteomics studies adopt a bottom-up proteomics strategy, in which proteins are enzymatically digested into peptides prior to LC-MS/MS analysis, and protein identities and abundances are subsequently inferred from the detected peptides. This approach reduces sample complexity and enables robust, high-throughput identification and quantification across large numbers of proteins.
In some experimental designs, stable isotope or isobaric labeling strategies, such as TMT or SILAC, may be introduced at this stage to enable multiplexed quantitative comparisons across multiple samples within a single LC-MS/MS experiment. (Learn more at: Label-based Protein Quantification Technology—iTRAQ, TMT, SILAC)
Peptide Separation and LC-MS/MS Acquisition
After digestion, peptides are separated by liquid chromatography to reduce sample complexity and improve the detection of low-abundance species. The separated peptides are then introduced into a tandem mass spectrometer, where their mass-to-charge (m/z) ratios are measured and selected ions are fragmented to generate sequence-informative MS/MS spectra.
Two data acquisition strategies are commonly used in discovery proteomics: data-dependent acquisition (DDA) and data-independent acquisition (DIA). In DDA, the mass spectrometer dynamically selects the most intense precursor ions for fragmentation in real time, a strategy that has historically dominated discovery proteomics workflows due to its technical maturity and robust identification performance.
However, although DDA has been widely used in discovery proteomics, its stochastic precursor selection can lead to missing values across runs, particularly for low- to medium-abundance peptides. This limitation becomes more pronounced as sample numbers increase. DIA addresses this challenge by systematically fragmenting all ions within predefined m/z windows, ensuring that peptides present in the sample are consistently sampled across runs. One widely used implementation of DIA is SWATH-MS (Sequential Window Acquisition of All Theoretical Mass Spectra), which acquires comprehensive fragment ion information across the entire mass range and enables reproducible, large-scale proteome quantification. As a result, DIA typically provides improved quantitative consistency and reproducibility, making it especially well suited for large-scale and cohort-based proteomics studies.
Protein Identification and Quantification
After peptide separation and LC-MS/MS acquisition, the next critical step in a discovery proteomics workflow is to convert raw spectral data into meaningful protein identifications and quantitative measurements. During this stage, the MS/MS spectra generated by the mass spectrometer are computationally matched to protein sequence databases using dedicated search algorithms. In this process, experimental spectra are compared with theoretical spectra derived from in silico digestion of protein sequence entries to find the best peptide matches, a process known as peptide–spectrum matching (PSM). To ensure the reliability of these matches, statistical scoring models and target-decoy approaches are applied to control the false discovery rate (FDR), which is typically set at ≤1% at both the peptide and protein levels to minimize incorrect identifications.
Once peptides are confidently identified, multiple PSMs are assembled into protein groups, considering shared and unique peptide evidence. Protein abundance can then be quantified using label-free quantification (LFQ), which compares peptide signal intensities across different runs, or labeling-based strategies such as isobaric tagging methods (e.g., TMT, SILAC), which enable multiplexed quantitative comparisons of multiple samples in the same experiment.
Together, these steps yield a quantitative protein matrix—a structured table of protein abundance values across all samples—that serves as the foundation for downstream statistical analysis, differential expression testing, and biological interpretation.
Data Analysis in Discovery Proteomics
Data analysis is where discovery proteomics delivers its greatest value, transforming large-scale protein measurements into biological insight.
Data Preprocessing and Normalization
Before downstream statistical analysis, quantitative proteomics data require careful preprocessing to reduce technical noise. Normalization, quality filtering, and appropriate handling of missing values are critical steps to ensure that observed differences represent true biological variation rather than artifacts introduced during sample preparation or mass spectrometry acquisition.
Differential Expression Analysis
Once the data are properly normalized, proteins are statistically compared across experimental groups to identify significant abundance changes. Differential expression analysis typically involves calculating fold changes, performing statistical hypothesis testing, and applying multiple-testing correction to control false discoveries.
Visualization methods such as volcano plots, heatmaps, and principal component analysis are often used to explore global trends, assess sample clustering, and highlight proteins of interest.
Functional and Pathway Interpretation
To move beyond lists of differentially expressed proteins, discovery proteomics data are commonly integrated with biological knowledge bases. Functional enrichment analyses map proteins to Gene Ontology terms, signaling pathways, and molecular networks, enabling researchers to interpret proteomic changes in a biological and mechanistic context.
This systems-level interpretation helps connect quantitative proteomics results to cellular processes, disease mechanisms, and therapeutic hypotheses.
Choosing Between DDA and DIA in Discovery Proteomics
Selecting an appropriate acquisition strategy is a key decision in discovery proteomics experiments. Data-dependent acquisition (DDA) has traditionally been favored for deep proteome identification and spectral library generation, benefiting from well-established workflows and extensive software support.
Data-independent acquisition (DIA), in contrast, systematically fragments all ions within defined m/z windows, enabling more consistent peptide sampling and improved quantitative completeness across runs. DIA-based discovery proteomics is therefore increasingly preferred for large-scale, cohort-based, and longitudinal studies where reproducibility and reduced missing values are critical.
The choice between DDA and DIA depends on study goals, sample complexity, cohort size, and available data analysis infrastructure. In practice, many workflows combine both approaches, using DDA for library generation and DIA for large-scale quantitative analysis.
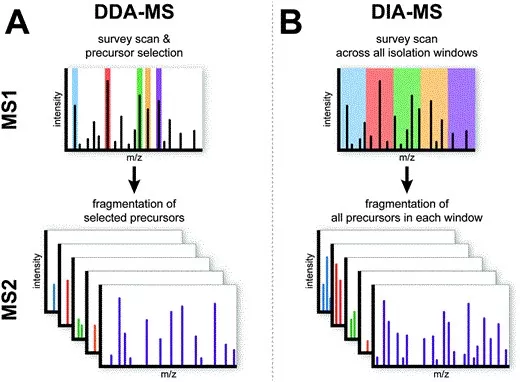
Comparison of Data-Dependent and Data-Independent Acquisition Strategies in Discovery Proteomics.
Image reproduced from Krasny and Huang, 2021, Molecular Omics, licensed under the Creative Commons Attribution 3.0 International License (CC BY 3.0).
Key Applications of Discovery Proteomics in Biomedical Research
Systems Biology and Pathway-Level Analysis
In systems biology, discovery proteomics is widely used to characterize global protein expression patterns, signaling pathways, and molecular interaction networks. Because proteins directly execute cellular functions, proteome-wide measurements provide functional insights that extend beyond genomic or transcriptomic data alone. Discovery proteomics datasets are frequently integrated with other omics layers, such as transcriptomics and metabolomics, to support network-based modeling of cellular processes and responses to genetic, environmental, or pharmacological perturbations.
Biomarker Discovery in Clinical Cohorts
Discovery proteomics is widely used in clinical research to identify proteins whose expression changes significantly between disease and control groups. These differentially expressed proteins can serve as candidate biomarkers for diagnosis, prognosis, or treatment stratification, and inform subsequent targeted validation.
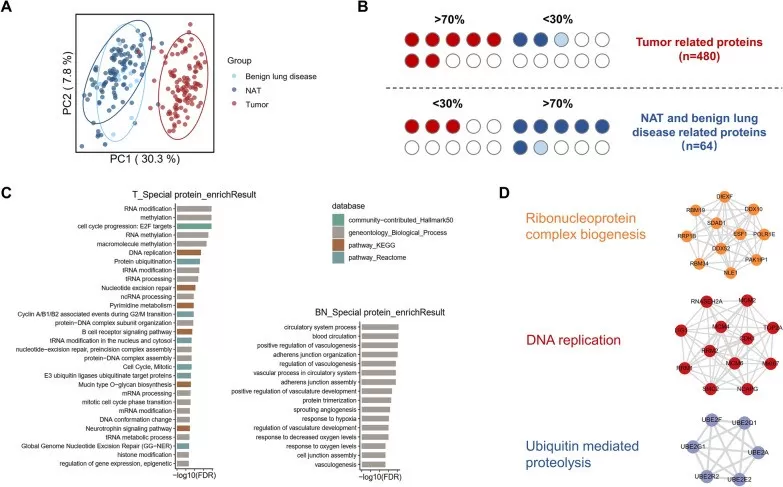
Discovery proteomics–based biomarker analysis in lung adenocarcinoma patients.
Image reproduced from Huang et al., Clinical Proteomics, 2024, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
In this example from a lung adenocarcinoma cohort, thousands of proteins were quantified and statistically analyzed to reveal proteins with significant differential abundance between patient groups, illustrating how discovery proteomics informs biomarker discovery and pathway insights.
Drug Discovery and Mechanism-of-Action Studies
In drug discovery and development, discovery proteomics supports target identification, pathway analysis, and mechanism-of-action studies. Proteome-wide profiling enables researchers to assess how compounds or genetic perturbations modulate protein expression and signaling networks at a systems level. Such analyses can reveal on-target and off-target effects, inform lead optimization, and contribute to early safety and efficacy assessments before progression to later-stage development.
Large-Scale and Cohort-Based Proteomics Studies
Advances in mass spectrometry acquisition strategies and data analysis workflows have enabled discovery proteomics to scale to large sample cohorts. In population-level and longitudinal studies, reproducible quantitative profiling across many samples allows researchers to investigate proteomic variability, disease heterogeneity, and temporal changes associated with disease progression or therapeutic intervention. These large-scale discovery proteomics datasets provide valuable resources for understanding complex disease biology and supporting precision medicine research.
Advantages and Limitations of Discovery Proteomics
Discovery proteomics provides a powerful framework for unbiased, proteome-wide analysis, enabling the identification and quantification of thousands of proteins without prior target selection. This global coverage makes it particularly valuable for exploratory studies, hypothesis generation, and pathway-level analysis, where the relevant proteins and biological mechanisms are not yet fully defined. Advances in mass spectrometry performance and data-independent acquisition (DIA) have further improved the reproducibility and quantitative consistency of discovery proteomics, supporting its application in large-scale and cohort-based studies.
At the same time, discovery proteomics faces inherent limitations. The wide dynamic range of protein abundance in biological samples can limit the detection of low-abundance proteins, and stochastic sampling in data-dependent acquisition (DDA) workflows may lead to missing values across runs. Although DIA reduces these issues, it introduces increased computational complexity and reliance on robust data analysis pipelines. In addition, protein inference from peptide-level data can be ambiguous for protein families with shared sequences, underscoring the need for careful experimental design and downstream validation.
Best Practices for High-Quality Discovery Proteomics
High-quality discovery proteomics relies not only on advanced mass spectrometry technologies, but also on rigorous experimental design, standardized data processing, and transparent reporting practices. Careful control of sample preparation, consistent LC–MS/MS acquisition settings, appropriate statistical analysis, and robust quality control are essential to ensure reliable and biologically meaningful results.
To promote reproducibility and data transparency, discovery proteomics studies often follow MIAPE (Minimum Information About a Proteomics Experiment) guidelines, which define the essential experimental and analytical details that should be reported when sharing proteomics data. These guidelines cover key aspects of a proteomics workflow, including sample origin, experimental design, mass spectrometry acquisition parameters, database searching strategies, and statistical analysis methods. Adherence to MIAPE standards enables accurate interpretation of results, facilitates independent validation, and supports reliable reuse of proteomics datasets in large-scale or comparative studies.
Together, these best practices help ensure that discovery proteomics (DIA proteomics and DDA proteomics) data are robust, reproducible, and suitable for downstream biological interpretation and translational applications.
Reference
1. Krasny, L., & Huang, P. H. (2021). Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology. Molecular omics, 17(1), 29–42. https://doi.org/10.1039/d0mo00072h
2. Huang, Y., Ma, S., Xu, J. Y., Qian, K., Wang, Y., Zhang, Y., Tan, M., & Xiao, T. (2024). Prognostic biomarker discovery based on proteome landscape of Chinese lung adenocarcinoma. Clinical proteomics, 21(1), 2. https://doi.org/10.1186/s12014-023-09449-2
Read more
- Proteomics Quality Control: A Practical Guide to Reliable, Reproducible Data
- Protein Complexes: What They Are and Why They Matter in Biomedical Research
- Blood Proteomics: Serum or Plasma – Which Should You Choose?
- Proteomics and Metabolomics/Lipidomics in Metabolic Disease Research: Insights and Applications
- Mass Spectrometry Acquisition Mode Showdown: DDA vs. DIA vs. MRM vs. PRM
- Proteomics Platform Showdown: MS-DIA vs. Olink vs. SomaScan
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


