FDR Control in Proteomics: Principles, Calculation Methods, and Threshold Selection
In bottom-up LC-MS/MS-based quantitative proteomics, accurately identifying peptides and proteins from vast amounts of mass spectrometry data presents a core challenge. False Discovery Rate (FDR) control is a statistical cornerstone to ensure the reliability of these identification results. This article systematically explores the necessity of FDR control, the principles and methods for FDR calculation based on the Target-Decoy strategy, the stepwise FDR calculation from peptides to proteins, and considerations for threshold selection. It also discusses the limitations of this strategy, providing researchers with a comprehensive and rigorous framework for data quality control.
1. The Importance of FDR Control in Proteomics
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) is capable of generating hundreds of thousands to millions of MS/MS spectra in a single analysis. In quantitative proteomics analysis, these spectra are matched with theoretical peptide sequences using database search algorithms to identify proteins. However, when performing such large-scale hypothesis testing (i.e., determining whether each spectrum match is valid), traditional p-value control methods (e.g., Bonferroni correction) can be overly conservative, leading to the loss of valuable biological signals. Without global error control, the number of false positives generated by random matching becomes unacceptably high, which severely distorts the biological conclusions drawn from the data.
Thus, there is a need for an error metric suitable for large-scale exploratory studies—False Discovery Rate (FDR). FDR is defined as the expected proportion of false positives among all accepted discoveries (e.g., identified peptides or proteins). By controlling FDR, we can report results with quantifiable confidence, such as “X proteins identified with FDR < 1%,” indicating that we expect no more than 1% of errors in the identified proteins.
2. Target-Decoy Strategy: A Reliable Approach for FDR Estimation
The Target-Decoy strategy is widely recognized as the most effective and reliable approach for estimating and controlling the False Discovery Rate (FDR) in proteomics. By incorporating both a target and a decoy database, this strategy allows for more accurate estimation of false positives and minimizes the risk of overestimating protein identifications.
2.1 Core Principles and Search Strategies for FDR Estimation
The Target-Decoy strategy is based on the assumption that the probability of obtaining false matches from the target database is the same as from the decoy database. The core idea is to construct a decoy database by reversing or scrambling the amino acid sequences of real proteins to create peptide sequences that are biologically implausible. Then, both the target and decoy databases are used to match experimental MS/MS spectra. There are two search strategies: separate search of target and decoy databases or combined target-decoy database search [1]. In the separate search, target and decoy databases are searched independently, and FDR is estimated using Kall’s method [2]. Each peptide spectrum has a best match from both the target and decoy databases. In the combined approach, a unified target-decoy database is searched, and peptides from both databases compete against each other for the best match. Each spectrum has one best score, either from the target or decoy database, but not both (Fig. 1).
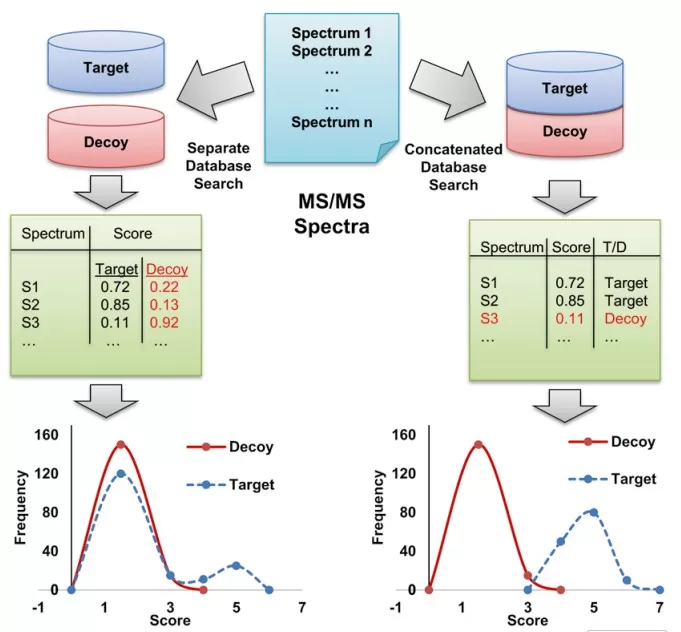
Fig. 1 two database search strategies. separate or combined database search. In separate search target and decoy databases are searched separately and FDR is estimated using Kall’s method . Each spectrum has one target and one decoy best score. In combined approach, one unified targetdecoy database is searched in which both TD peptides compete with each other. Each spectrum has one best score, either from target or decoy but not both. This also changes the score distributions [1]
2.2 FDR Calculation Methodologies and Best Practices
The general process for calculating FDR is as follows (Fig. 2):
1) Rank all peptide spectra from both the target and decoy databases by score, from best to worst.
2) For each target score threshold, calculate the number of decoy (D) and target (T) spectra above that threshold, as well as the number of decoy and target peptides above the threshold.
3) FDR is then calculated using the following formula (for more detailed methods, refer to [1]).
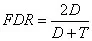
Where D represents the number of decoy spectra above the threshold, and T represents the number of target spectra above the threshold. Protein-level FDR calculations follow a similar process to the peptide-level calculations described above.
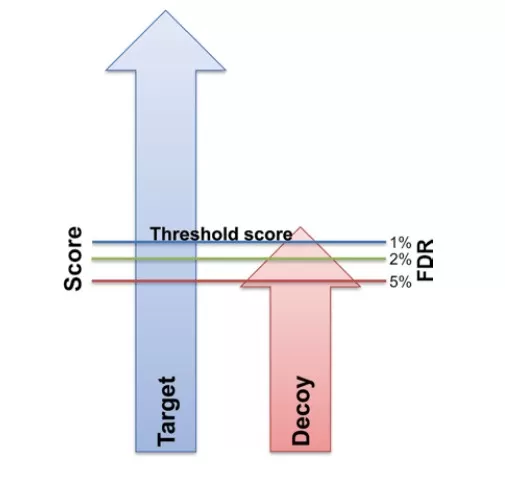
Fig. 2: Illustration of the Target-Decoy false positive rate evaluation strategy
3. Step-by-Step FDR Calculation: From Peptides to Proteins
Proteomics identification is a multi-step process, and FDR control must be applied throughout each stage.
3.1 Peptide-Level FDR Control
Analysis typically starts by controlling FDR at the peptide-spectrum matching level (commonly set at 1%), resulting in a high-confidence list of peptide identifications.
Key decision: Handling shared peptides. "Shared peptides" are those that exist in multiple proteins (e.g., homologous proteins or isoforms). Research has shown that including shared peptides during the protein inference step can lead to substantial distortion in FDR estimates, making them overly lenient. The best practice is to use "unique peptides," which can uniquely map to a single protein or protein group, to ensure the accuracy of FDR estimates in subsequent protein inference.
3.2 Protein-Level FDR Control
Directly applying peptide-level FDR to protein-level identification presents a major issue: in large-scale datasets, as real proteins are repeatedly identified, the pool of remaining target protein sequences that could be matched by false positives shrinks, while the decoy sequence pool remains "full." This results in an overestimation of protein-level false positives in the classical Target-Decoy strategy.
To address this, Savitski et al. proposed the innovative "Picked" Target-Decoy strategy [3]. In this method, each target protein sequence is paired with its corresponding decoy sequence. During protein inference, for each pair, only the higher-scoring sequence is "picked" as the representative (whether from the target or decoy). If the target has the higher score, it is counted as a target protein identification; if the decoy has the higher score, it is counted as a decoy protein identification (Fig. 3). This symmetrical and unbiased approach prevents the overrepresentation of decoy proteins in large datasets, resulting in stable and reliable protein-level FDR estimates.
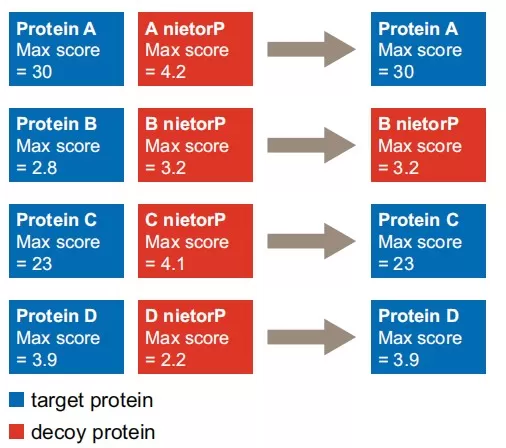
Fig. 3: Illustration of the "Picked" strategy for protein-level FDR control
3.3 Protein Score Calculation and Inference Methods
Once unique peptides and the "Picked" strategy are applied, a summary score for each candidate protein group must be calculated. Several methods for this have been compared:
1) Best peptide score: The highest score from all associated unique peptides of the protein.
2) Peptide posterior error probability (PEP) product: The product of the posterior error probabilities of all associated unique peptides.
3) Fisher method: The combination of p-values for all associated unique peptides.
4) Two-peptide rule: Requires at least two unique peptides to support a protein.
In large, complex datasets, the "best peptide score" method has demonstrated superior performance and robustness, identifying the truest proteins while maintaining effective FDR control.
4. Choosing the Optimal FDR Threshold for Accurate Identification
In proteomics, an empirically accepted standard is to control identification results at 1% FDR. This threshold is statistically significant and widely agreed upon in the industry. A 1% FDR means that for every 100 reported identifications, only one is expected to be false, which is considered a high-confidence result. Why not lower? As FDR decreases, more true signals are filtered out. At 0.1% FDR, many marginal but true peptides may be discarded, leading to a significant loss in sensitivity. At 5% FDR, false positives increase substantially, compromising specificity. A 1% threshold strikes a good balance between sensitivity and specificity, allowing researchers to identify more proteins while keeping the overall error rate low. In fact, the 1% FDR threshold was derived from early large-scale proteomics studies, which found that the curve's inflection point commonly occurred around 1%, where further reduction in FDR resulted in a sharp decline in identification numbers with only a slight reduction in false positives [4]. As a result, 1% FDR has become the default standard.
However, threshold selection is not absolute. Researchers should adjust the FDR threshold based on specific experimental goals: exploratory discovery studies may accept a slightly higher FDR (e.g., 5%) to maximize the scope of discoveries, but more resources should be invested in follow-up validation. For targeted validation or clinical biomarker studies, stricter thresholds (e.g., 0.1% or lower) should be used to ensure the reliability of each identified target.
5. Final Considerations for Reliable FDR Estimation in Proteomics
The Target-Decoy strategy offers a simple yet powerful method for controlling false positives in mass spectrometry-based proteomics, but it is not without limitations. When applying this strategy, it is essential to keep its statistical assumptions in mind and adjust for special cases. By combining rigorous multi-level validation (such as entrapment testing), reasonable data filtering strategies, and correction for potential biases, researchers can trust the FDR estimates from the Target-Decoy strategy in most cases. However, when operating beyond its applicability limits (e.g., extremely low signal data or complex homologous backgrounds), caution is needed, and supplementary methods should be incorporated as necessary. Maintaining a critical understanding of FDR control strategies is key to ensuring robust and reliable scientific conclusions.
References
1. Aggarwal, S. and A.K. Yadav, False Discovery Rate Estimation in Proteomics. Methods Mol Biol, 2016. 1362: p. 119-28.
2. Kall, L., et al., Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J Proteome Res, 2008. 7(1): p. 29-34.
3. Savitski, M.M., et al., A Scalable Approach for Protein False Discovery Rate Estimation in Large Proteomic Data Sets. Mol Cell Proteomics, 2015. 14(9): p. 2394-404.
4. Wang, G., et al., Decoy methods for assessing false positives and false discovery rates in shotgun proteomics. Anal Chem, 2009. 81(1): p. 146-59.
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


