Mass Spectrometry Imaging (MSI): A Comprehensive Guide to Techniques, Applications, and Future Trends
In the rapidly evolving landscape of multi-omics, the ability to visualize "where" molecules are located is as critical as knowing "what" they are. Traditional liquid chromatography-mass spectrometry (LC-MS) remains a powerhouse for quantification, but it requires tissue homogenization, which inherently destroys the spatial architecture of the sample.
Mass Spectrometry Imaging (MSI) is revolutionizing the way scientists visualize and analyze the molecular composition of biological tissues. By combining the power of mass spectrometry with imaging technologies, MSI enables researchers to obtain high-resolution, spatially resolved maps of metabolites, proteins, lipids, and other biomolecules within tissue samples. This cutting-edge technology is paving the way for breakthroughs in fields such as biomarker discovery, oncology, and PK/PD studies.
In this article, we will provide a comprehensive overview of MSI, covering its fundamental principles, types, workflows, challenges, applications, and future trends.
1. What is Mass Spectrometry Imaging (MSI)?
Mass Spectrometry Imaging (MSI) is a technique that allows for the direct visualization of the distribution of molecules within a tissue sample by combining mass spectrometry with imaging. MSI generates molecular maps that show how specific compounds are spatially distributed across tissue sections. Unlike immunohistochemistry (IHC) or fluorescence microscopy, MSI is a "label-free" molecular imaging technique, does not require specific antibodies or fluorescent tags, making it an unbiased tool for metabolite mapping.
MSI works by ionizing molecules from the surface of a tissue sample and measuring the mass-to-charge ratios of the ions. These measurements are used to create a spatial map that indicates where specific compounds are located within the sample. The molecular distribution is represented in color-coded images, providing a clear picture of the biochemical composition of the tissue.
The main components of MSI include:
- Ionization method: A technique such as MALDI, DESI, or SIMS is used to ionize the molecules.
- Mass spectrometry: After ionization, the molecules are analyzed in a mass spectrometer to determine their mass-to-charge ratios.
- Data processing: The resulting data is processed to create an image that visualizes the spatial distribution of the compounds.
MSI allows researchers to observe the molecular composition of tissues in situ, preserving the spatial context, which is crucial for understanding complex biological processes.
2. Types of Mass Spectrometry Imaging Techniques: MALDI, DESI, and SIMS
Several MSI techniques are used depending on the type of sample, the molecules of interest, and the desired resolution. Below are the most common types:
 techniques based on ionizatio_1768368394_WNo_850d584.webp)
Classification of in situ mass spectrometry imaging (MSI) techniques based on ionization methods
Image reproduced from Shen et al., 2025, Rapid Communications in Mass Spectrometry
2.1 MALDI-MS Imaging (Matrix-Assisted Laser Desorption/Ionization)
MALDI‑MS Imaging is one of the most widely used modalities within Mass Spectrometry Imaging (MSI), prized for its ability to sensitively visualize a broad range of biomolecules directly from thin tissue sections. The technique relies on applying a chemical matrix—a small organic molecule that co‑crystallizes with the analyte—across the sample surface. Upon irradiation with a UV or IR laser, the matrix absorbs the energy and facilitates soft ionization of intact molecules, enabling the detection of proteins, lipids, peptides, and metabolites with minimal fragmentation. This soft ionization mechanism gives MALDI a competitive edge: unlike DESI or SIMS, MALDI can seamlessly transition from imaging small metabolites to large proteins (>100 kDa), offering a superior balance of spatial resolution (typically ~5–20 µm) and chemical sensitivity. However, the achievable resolution and signal quality are strongly influenced by the method of matrix application—for example, sublimated versus sprayed matrices produce different crystal sizes and ionization efficiencies, which directly impact image quality and reproducibility.
2.2 DESI-MS Imaging (Desorption Electrospray Ionization)
DESI‑MS Imaging enables ambient mass spectrometry imaging under room temperature and atmospheric pressure, eliminating the need for vacuum systems or extensive sample preparation, which greatly simplifies the workflow compared to traditional MSI methods like MALDI and SIMS. In DESI, a charged solvent spray impacts the tissue surface to form a thin liquid film that extracts analytes, which are then desorbed and ionized into the mass spectrometer for analysis. This ambient, “plug‑and‑play” ionization is ideal for small molecule metabolites and polar lipids, with minimal chemical background from matrices, making DESI particularly suitable for clinical environments and spatial metabolomics studies where quick, direct tissue analysis is needed. Additionally, because DESI is essentially non‑destructive, the same tissue section can subsequently be used for histological staining or immunohistochemistry, facilitating precise co‑registration of molecular and morphological information in multi‑modal workflows.
2.3 SIMS-MS Imaging (Secondary Ion Mass Spectrometry)
SIMS uses a high-energy primary ion beam to bombard the tissue surface, causing the release of secondary ions that are then analyzed by the mass spectrometer. This direct sputtering mechanism gives SIMS exceptional spatial resolution, often down to the submicron and even nanoscale level (e.g., ~25 nm with advanced cluster beams), which allows detailed chemical mapping at cellular and subcellular scales—far beyond the typical resolution of MALDI or DESI. SIMS is often described as a “harder” ionization technique compared with MALDI and DESI because the energetic ion impact can lead to extensive fragmentation of larger biomolecules. As a result, traditional SIMS is particularly strong for imaging elements, isotopes, small molecular fragments, lipids, and metabolites with high lateral precision, but less effective for intact large proteins.
Comparative Analysis of MSI Technologies (MALDI vs. DESI vs. SIMS)
|
Feature |
MALDI-MSI |
DESI-MSI |
SIMS-MSI |
|
Ionization Source |
Matrix-Assisted Laser Desorption/Ionization |
Desorption Electrospray Ionization |
Secondary Ion Mass Spectrometry |
|
Atmospheric Requirements |
High Vacuum or Ambient (AP-MALDI) |
Ambient (Atmospheric Pressure) |
High Vacuum |
|
Spatial Resolution |
High (1–20 µm; Single-cell capable) |
Moderate to High (10–100 µm) |
Ultra-High (50 nm – 1 µm; Subcellular) |
|
Sensitivity |
High (Excellent for peptides/proteins) |
Moderate (Optimized for small molecules) |
High (Limited to elements/fragments) |
|
Mass Range |
Wide (100 to >100,000 Da) |
Narrow to Moderate (<2,000 Da) |
Narrow (<1,000 Da due to fragmentation) |
|
Matrix Requirement |
Essential (Requires chemical matrix) |
Matrix-free (Direct solvent spray) |
Matrix-free (Direct ion beam) |
|
Ionization Hardness |
Soft (Intact molecular ions) |
Soft (Non-destructive) |
Hard (Significant fragmentation) |
|
Sample Preparation |
Intensive (Sectioning, matrix coating) |
Minimal (Direct analysis) |
Intensive (Dehydration, ultra-flat surface) |
|
Acquisition Speed |
Fast (High-frequency lasers) |
Fast (Real-time monitoring) |
Slow (High-resolution rastering) |
|
Primary Applications |
Comprehensive metabolite imaging, tissue profiling in animals/plants |
In situ tissue imaging, plant research, clinical applications |
elemental mapping, nanoscale surface analysis |
3. How Does Mass Spectrometry Imaging Work? The General Workflow
Mass Spectrometry Imaging (MSI) follows a structured, multi-step workflow that transforms biological tissue into spatially resolved molecular maps, enabling high-resolution spatial metabolomics and multimolecular analysis. Each stage is critical to ensure data quality, reproducibility, and accurate localization of molecules within tissue contexts.
Step 1: Sample Preparation
MSI begins with preserving and sectioning tissue into thin slices (typically 5–20 µm) to retain native molecular distribution. For vacuum‑based methods like MALDI and SIMS, sections are usually mounted on conductive substrates to support stable ionization under vacuum. In contrast, DESI operates at ambient pressure and does not require conductive slides, allowing simpler sample mounting and minimal pretreatment.
Step 2: Ionization
After sectioning, molecules on the tissue surface are ionized using a technique suited to the target chemistry and resolution: a laser for MALDI, a charged solvent spray for DESI, or an ion beam for SIMS. The chosen ionization method directly influences the types of compounds detected and the achievable spatial resolution, making this a decisive step in the workflow.
Step 3: Mass Spectrometry Analysis
The generated ions are transferred into a mass spectrometer, where they are separated based on their mass-to-charge (m/z) ratios. Each pixel of the tissue scan yields a full mass spectrum that reflects the local molecular composition. High-resolution analyzers or time-of-flight detectors help distinguish between closely spaced m/z values and capture a broad range of molecular species.
Step 4: Data Processing and Image Reconstruction
Once raw spectra are collected across the tissue grid, specialized software compiles the data into spatially mapped images. Each m/z value can be visualized as a colored heat map, enabling direct comparison of molecular distributions within the tissue. Computational tools also perform peak picking, normalization, and alignment to reduce noise and improve interpretability.
Step 5: Interpretation and Analysis
The reconstructed images are then analyzed to reveal biological insights—identifying patterns, co-localization of molecules, and differences between sample groups (e.g., healthy vs. diseased). Quantitative and comparative analyses can uncover potential biomarkers, elucidate metabolic pathways, or support spatial omics integration with histological and genomic data for comprehensive tissue profiling.
 workflow._1768368737_WNo_849d578.webp)
Mass spectrometry imaging (MSI) workflow.
Image reproduced from Porta et al., 2018, Molecular Imaging and Biology, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
4. Popular Applications of Mass Spectrometry Imaging
Mass Spectrometry Imaging (MSI) has rapidly become a versatile tool in biomedical research and translational applications because it can visualize the spatial distribution of hundreds to thousands of endogenous and exogenous molecules simultaneously without labeling. This unique capability has enabled breakthroughs in understanding disease biology, discovering biomarkers, studying drug distribution, and mapping complex tissue biochemistry in situ.
4.1 Disease Mechanism Mapping
MSI is widely used to dissect the biochemical mechanisms underlying diseases by mapping spatial molecular changes across diseased and normal tissues. For example, spatial metabolomics approaches based on MSI have revealed heterogeneous metabolic signatures within tumor microenvironments, providing insights into tumor metabolism, cell signaling, and microenvironment interactions that are not accessible with bulk analyses. This spatial context helps elucidate how metabolic dysregulation contributes to disease progression and cellular heterogeneity in conditions such as cancer and neurological disorders.
4.2 Biomarker Discovery in Complex Tissues
By directly comparing the spatial distribution of molecular species between healthy and diseased sections, MSI enables the identification of potential biomarkers that correlate with pathology. In cancer metabolomics, for instance, MSI can differentiate metabolic heterogeneity within tumors, highlighting metabolites and lipid species that associate with tumor grade or response to therapy, thus aiding both diagnostic stratification and hypothesis generation for new therapeutic targets.
4.3 Drug Distribution and Pharmacology
MSI has become an essential tool in drug development and pharmacology research because it can map the spatial distribution of drugs, metabolites, and endogenous molecules simultaneously in tissue sections. Unlike traditional bioanalytical methods that provide average concentrations, MSI shows how drug compounds and their metabolites localize within organs or tumor regions in situ, helping researchers evaluate drug penetration, retention, and metabolic transformation directly in biological tissue contexts.
4.4 Neuroscience and Brain Mapping
In neuroscience research, MSI enables high‑resolution mapping of neurotransmitters, small metabolites, lipids, and metabolic alterations across brain regions, offering a spatially resolved molecular perspective on brain function and pathology. For example, advanced ambient MSI strategies have been used to characterize metabolic profiles of mouse brain microregions, revealing distinct distributions of metabolites linked to neuronal activity and regional specialization. This spatial information can deepen understanding of neurological disease mechanisms and brain metabolome organization.
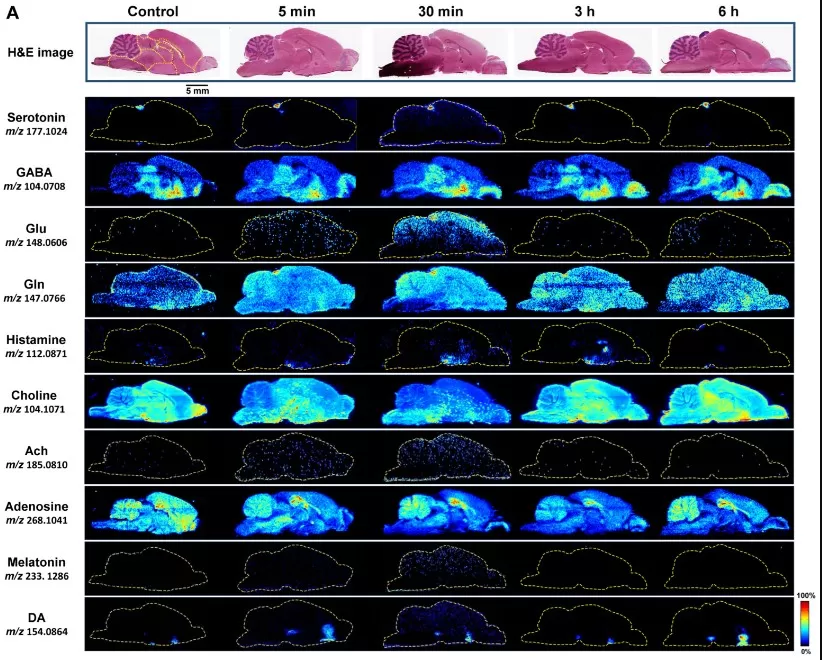
Spatiotemporal distribution of neurotransmitters in the brain following oral 1,8-cineole administration. Representative H&E-stained sections and corresponding AFADESI-MSI maps at 5 min, 30 min, 3 h, and 6 h post-dose.
Image reproduced from Wu et al., 2025, Metabolites, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
5. Future Trends and Developments in Mass Spectrometry Imaging
Mass Spectrometry Imaging (MSI) continues to evolve rapidly, driven by advances in instrument performance, data analytics, and integration with other biological technologies. The following are key trends shaping the future of MSI:
1) Higher Spatial Resolution
MSI techniques are advancing toward cellular and subcellular spatial resolution, enabling detailed mapping of metabolites and molecular features at previously inaccessible scales. This trend is especially important for single‑cell spatial metabolomics and nanoscale biochemical imaging.
2) Integration with Multi‑Omics
MSI is increasingly combined with other omics layers (e.g., genomics, transcriptomics, proteomics) to build spatially resolved multi‑omics maps, offering deeper mechanistic insights than any single modality.
3) Improved Quantification and Standardization
Efforts are focused on enhancing quantitative accuracy and reproducibility across labs, addressing current challenges in signal variability and calibration for more reliable comparative studies.
4) High‑Throughput and Automation
To meet the needs of large‑scale and clinical studies, MSI workflows are moving toward automation and faster acquisition, reducing bottlenecks in sample processing and data collection.
5) Advanced Computational Tools and AI
AI and deep learning methods are being developed to improve MSI data processing, feature extraction, and pattern recognition, making it easier to interpret complex spatial datasets.
Reference
1. Porta Siegel, T., Hamm, G., Bunch, J. et al. Mass Spectrometry Imaging and Integration with Other Imaging Modalities for Greater Molecular Understanding of Biological Tissues. Mol Imaging Biol 20, 888–901 (2018). https://doi.org/10.1007/s11307-018-1267-y
2. Shen, X., Zhang, F., Tang, C., Soković, M., Mišić, D., Xu, H., Ye, Y. and Liu, J. (2025), Advances in Sampling and Analytical Techniques for Single-Cell Metabolomics: Exploring Cellular Heterogeneity. Rapid Commun Mass Spectrom, 39: e10045. https://doi.org/10.1002/rcm.10045
3. Wu J, Mu Q, Qi J, Bao H, Sa C. Ambient Mass Spectrometry Imaging Reveals Spatiotemporal Brain Distribution and Neurotransmitter Modulation by 1,8-Cineole: An Epoxy Monoterpene in Mongolian Medicine Sugmel-3 . Metabolites. 2025; 15(9):631. https://doi.org/10.3390/metabo15090631
Read more
- Spatial Metabolomics: Transforming Biomedical and Agricultural Research
- Spatial Metabolomics Explained: How It Works and Its Role in Cancer Research
- MALDI, DESI, or SIMS? How to Choose the Best MSI Techniques for Spatial Metabolomics
- How to Prepare Samples for Spatial Metabolomics: The Essential Guide You Need
- Unlocking Precision in Spatial Metabolomics: Essential Detection Parameters for Cutting-Edge Research
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


