Nucleosides: Structure, Functions, Biomarkers & LC–MS/MS Profiling
Nucleosides are more than just chemistry jargon – they’re fundamental to life and health. These molecules, composed of a sugar and a nitrogenous base, form the core of our genetic material and fuel many biological processes. Understanding nucleosides can illuminate how our cells replicate, how certain medicines work, and even how clues in our urine might reveal disease. In this comprehensive guide, we’ll explore what nucleosides are, how they were discovered, how they differ from nucleotides, and why they matter for everything from plant growth to human health. By the end, you’ll see how these small compounds influence daily life and how cutting-edge metabolomics services can profile nucleosides to advance research and wellness.
Overview:
What Are Nucleosides? Discovery, Structure, and Types
Nucleoside vs Nucleotide: Key Structural & Functional Differences
Biosynthesis: Making and Moving Nucleosides
Human Nucleoside Metabolic Pathways
Nucleoside Salvage Pathway & Degradation
Biological Functions of Nucleosides: Precursors, Signals, and Pharmacology
Nucleoside Analog Drugs: Chemotherapy’s Double-Edged Swords
Urinary Nucleosides as Non-Invasive Biomarkers: The Story in Our Pee
Nucleosides in Plants: Cytokinin Signaling, Purine Salvage, and Stress-Responsive Growth
Daily Life with Nucleosides: Supplements, Umami, Caffeine, and mRNA Vaccines
How to Measure Nucleosides: LC–MS/MS and Other Analytical Methods
What Are Nucleosides? Discovery, Structure, and Types
What Are Nucleosides: Nucleosides are versatile molecular “currencies” that move between cells and pathways. They act as mobile precursors for DNA/RNA synthesis (rapidly phosphorylated in salvage pathways), signaling cues (e.g., adenosine tuning blood flow, neural activity, and immunity), and diagnostic readouts (modified nucleosides accumulate in urine as non-invasive biomarkers). Medicinal chemistry also repurposes them as drug scaffolds—many frontline antivirals and chemotherapeutics are nucleoside analogs activated inside cells. Diet and microbiome influence circulating nucleosides, linking nutrition and metabolism to cellular proliferation and tissue repair.
_1756696437_WNo_271d293.webp)
The Discovery of Nucleosides: Nucleosides emerged on scientists’ radar in the late 19th and early 20th centuries during the quest to understand DNA/RNA. Pioneering biochemists like Albrecht Kossel identified component bases (adenine, cytosine, guanine, thymine, uracil) in nucleic acids, and Phoebus Levene later characterized the sugar-base units. By the 1930s, the term “nucleoside” was coined to describe a base linked to a sugar (ribose or deoxyribose), distinguished from “nucleotide” which also includes phosphate. This foundational work set the stage for Watson and Crick’s DNA model, where nucleosides are the building blocks of genetic strands.
Structure in Detail: A nucleoside is a nitrogenous base (purine: adenine/guanine; pyrimidine: cytosine/thymine/uracil) linked via an N-glycosidic bond to a five-carbon sugar—ribose in RNA nucleosides (adenosine, guanosine, cytidine, uridine) or 2′-deoxyribose in DNA counterparts (e.g., deoxyadenosine). Lacking phosphate, nucleosides are smaller and more membrane-permeable than nucleotides. Subtle alterations to the base or sugar (fluoro, azido, deoxy substitutions, etc.) dramatically change biological behavior, a principle leveraged to create nucleoside analog drugs and probes.
Nucleoside vs Nucleotide: Key Structural & Functional Differences
It’s easy to mix up nucleosides and nucleotides – their names sound similar and both relate to DNA/RNA. So how are nucleosides different from nucleotides? The key is the phosphate.
Nucleoside: Sugar + Base. No phosphate. Think of a nucleoside as a “letter” (base) with an attachment (sugar).
Nucleotide: Sugar + Base + Phosphate group(s). With one phosphate it’s a nucleoside monophosphate (e.g. AMP), with two or three it’s di- or triphosphate (ADP/ATP). Nucleotides are like the “letter” with a handle attached (phosphate) – the handle is what links letters into chains (DNA/RNA) and carries energy.
Structural Difference: The presence of phosphate in nucleotides gives them a negative charge and higher molecular weight. This means nucleotides are less able to cross cell membranes freely than nucleosides. Nucleosides, being uncharged, often serve as transport forms in the body that can slip through cell membranes or be carried by transporters.
Functional Difference: Nucleotides are the workhorses – they polymerize to form DNA/RNA strands and act as cellular energy currency (ATP/GTP). Nucleosides, on their own, don’t form genetic polymers. Instead, they often act as precursors or signals. For instance, cells import nucleosides and then add phosphate back to recycle them into nucleotides (salvage pathways). Some nucleosides like adenosine also have independent signaling roles, binding to receptors to modulate physiological responses (e.g. heart rate, inflammation).
In summary, nucleosides lack phosphate, whereas nucleotides have it, explaining differences in function and localization. The table below highlights these distinctions:
|
Feature |
Nucleoside |
Nucleotide |
|
Composition |
Base + Sugar (ribose or deoxyribose) |
Base + Sugar + Phosphate (one or more) |
|
Examples |
Adenosine, Cytidine, Thymidine, Guanosine |
AMP/ADP/ATP, CTP, dGTP, UMP, etc. |
|
Charge |
Neutral (no phosphate) |
Negative (due to phosphate) |
|
Role |
Precursors for nucleotides; signaling (e.g. adenosine) |
Building blocks of DNA/RNA; energy carriers (ATP) |
|
Ability to Polymerize |
Cannot form DNA/RNA chains directly |
Can polymerize (phosphate forms the backbone link) |
|
Transport |
Can cross cell membranes (via nucleoside transporters) |
Need to be synthesized inside cells or dephosphorylated to cross membranes |
Understanding this difference sets the stage for appreciating how nucleosides move around the body and why cells use special pathways to handle them.
Biosynthesis: Making and Moving Nucleosides
Nucleosides arise as part of nucleotide metabolism. Cells build nucleotides de novo from amino acids, CO₂, and one-carbon units, then interconvert them with nucleosides by removing or adding phosphate groups as demand shifts. A classic example is stress- or damage-triggered breakdown of extracellular ATP through ectonucleotidases (CD39/CD73) to adenosine, a signaling nucleoside that modulates vascular tone and immunity via surface receptors. Within cells, transient dephosphorylation also buffers nucleotide pools and facilitates export or reuse. The net effect is a dynamic, compartment-spanning circuit: nucleotides are synthesized when needed, nucleosides appear as transportable or signaling forms, and rapid interconversion maintains balance without committing to any specific pathway prematurely.
Human Nucleoside Metabolic Pathways
Human nucleoside metabolism integrates membrane transport, intracellular interconversion, and organ-level handling. Circulating adenosine, guanosine, uridine, and deoxyribonucleosides enter cells via equilibrative (ENT1–4; SLC29) and concentrative transporters (CNT1–3; SLC28), matching uptake to tissue demand. Inside cells, kinases rapidly funnel nucleosides to nucleotides for DNA/RNA synthesis or energy coupling; alternative routes channel bases and ribose-1-phosphate into central carbon pathways. Organ roles are distinct: the intestine supplies dietary nucleosides; the liver shapes systemic pools and first-pass metabolism; the kidney reclaims or eliminates filtered species; immune, brain, and muscle tissues tune uptake and salvage to function. Mitochondria maintain mtDNA via dedicated deoxynucleoside salvage. Together these fluxes preserve nucleotide homeostasis and couple metabolism to perfusion, inflammation, hypoxia responses, and cell-cycle status.
Nucleoside Salvage Pathway & Degradation
Salvage minimizes ATP cost by recycling nucleosides rather than rebuilding nucleotides de novo. Adenosine kinase (ADK), deoxycytidine kinase (dCK), thymidine kinase 1 (TK1), and uridine–cytidine kinases (UCK1/2) re-phosphorylate ribo- and deoxyribonucleosides to replenish DNA/RNA precursors. When supply exceeds demand, nucleoside phosphorylases (PNP, UPP1/2) and deaminases (ADA, CDA, GDA) convert nucleosides to bases plus ribose-1-phosphate that feed the non-oxidative pentose phosphate pathway and glycolysis. Downstream, purines are oxidized (xanthine oxidoreductase) to uric acid for renal excretion, whereas pyrimidines are reduced (DPYD) and further processed (DPYS/UPB1) to β-alanine or β-aminoisobutyrate, CO₂, and NH₃. Highly proliferative tissues lean on salvage to sustain synthesis; quiescent tissues favor degradation to balance pools and energy. Notably, many modified nucleosides (e.g., pseudouridine, 1-methyladenosine) resist salvage and are excreted in urine, creating non-invasive readouts of RNA turnover.
Biological Functions of Nucleosides: Precursors, Signals, and Pharmacology
Nucleosides serve five compact roles: (i) Precursors—phosphorylation supplies (d)NTPs for DNA/RNA synthesis during replication and repair; (ii) Metabolic support—uridine phosphorolysis generates ribose-1-phosphate to sustain central carbon flux and NADPH when glucose is limited; (iii) Signals—adenosine derived from AMP engages A1/A2A/A2B/A3 receptors to tune perfusion, neuronal excitability and immune tone; (iv) Cofactor scaffolds—adenosine moieties sit in ATP, NAD(P)H and CoA, tying nucleoside status to energy/redox; (v) Pharmacologic gateways—antivirals and chemotherapies are nucleoside analogs activated by cellular kinases. Together these functions link nucleoside availability to proliferation, stress adaptation, inflammation and therapy response—while avoiding overlap with the overview (transport) and biomarker sections (urinary panels).
Nucleoside Analog Drugs: Chemotherapy’s Double-Edged Swords
Nucleoside analogs are cornerstone scaffolds in modern drug R&D, powering antivirals and anticancer agents. Their pharmacology hinges on native uptake (ENT/CNT transporters) and stepwise phosphorylation by host or viral kinases to active mono/di/tri-phosphates. Once engaged, they terminate DNA/RNA strands, miscode bases, inhibit key enzymes (e.g., ribonucleotide reductase, thymidylate synthase), or trap DNA methyltransferases to reset epigenetic programs. Selectivity often derives from pathogen- or tumor-biased activation and polymerase preference. Resistance maps to transport (↓hENT1/CNT), activation (↓dCK/TK), deactivation (↑CDA, 5′-nucleotidases), and target repair, making these same nodes actionable biomarkers. Medicinal chemistry tunes base/sugar stereochemistry and fluorination, while “masked phosphate” ProTide prodrugs bypass the monophosphate bottleneck and improve delivery. Safety (notably myelosuppression), renal/hepatic handling, and combination strategies anchor development, as next-gen, transporter-agnostic and tumor-targeted designs push nucleoside therapeutics toward genuinely precise, durable regimens.
A recent case study illustrates both power and limits in the clinic. In pancreatic ductal adenocarcinoma (PDAC), gemcitabine—a deoxycytidine analog—often fails due to acquired resistance. A 2021 Frontiers in Oncology study identified the ketolysis enzyme OXCT1 as a driver of gemcitabine resistance via NF-κB activation: tumors and cell lines with high OXCT1 were less apoptotic under gemcitabine, and patients with elevated OXCT1 showed shorter relapse-free survival. Mechanistically, OXCT1 increased phosphorylation of IKKβ/IκBα/P65 and nuclear p-P65. Functionally, the NF-κB inhibitor BAY 11-7082 restored gemcitabine sensitivity in vitro and suppressed growth of OXCT1-overexpressing xenografts in vivo, pointing to a combination strategy (gemcitabine + NF-κB blockade) for OXCT1-high PDAC. Together, these data underscore how nucleoside analog efficacy hinges on cellular pathways beyond transport and phosphorylation, and how pathway-matched combinations can overcome resistance.
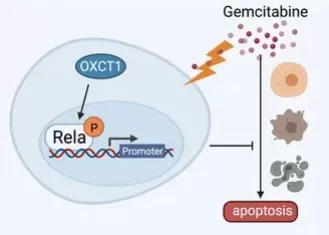
OXCT1-NF-κB–mediated gemcitabine resistance in PDAC.
Source: Dong J, Li H, et al. “OXCT1 Enhances Gemcitabine Resistance Through NF-κB Pathway in Pancreatic Ductal Adenocarcinoma,” Frontiers in Oncology (2021). Figure 6. Licensed under CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/). No endorsement implied.
Urinary Nucleosides as Non-Invasive Biomarkers: The Story in Our Pee
Imagine if a simple urine test could reveal if you have cancer or how your body is responding to treatment. It’s becoming reality, thanks to nucleosides. Modified nucleosides (like methylated versions) are constantly released into blood and filtered by the kidneys into urine. They come from the turnover of our RNAs—especially tRNA and rRNA, which contain many modified bases. In healthy people, these modified nucleosides are produced and excreted at baseline levels. But in diseases like cancer, the cellular turnover and RNA production are often ramped up, leading to elevated levels of certain nucleosides in urine.
A PLOS ONE study quantified six urinary nucleosides in 149 participants via HPLC–MS/MS. A six-nucleoside panel achieved 68% sensitivity for colon cancer with ≤2% false positives; combining the panel with serum CEA increased sensitivity to 85%, illustrating how urine nucleoside profiling can sharpen early detection and monitoring.
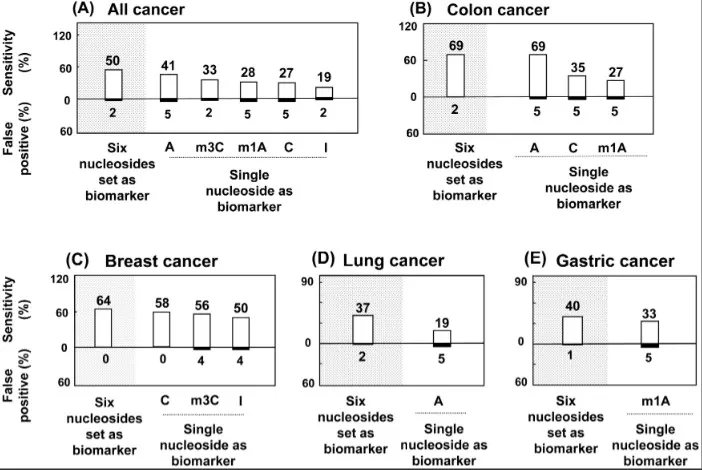
Diagnostic sensitivity of urinary nucleoside panels across cancers.
Source: Hsu W-Y et al. ‘Urinary Nucleosides as Biomarkers of Breast, Colon, Lung, and Gastric Cancer in Taiwanese,’ PLOS ONE (2013) 8(12): e81701. Figure 3A. Licensed under CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/). No endorsement implied.
Nucleosides in Plants: Cytokinin Signaling, Purine Salvage, and Stress-Responsive Growth
Plants deploy nucleosides far beyond genetics. As hormones, cytokinin ribosides (e.g., trans-zeatin riboside) move through xylem to coordinate shoot growth and stress responses, aided by dedicated transporters (ENTs, PUPs) and apoplastic nucleosidases that toggle between transport (riboside) and active base forms. As nutrient currency, purine salvage conserves nitrogen: nucleic acids and ATP catabolites are recycled, while surplus purines are funneled via xanthosine→xanthine into ureides (allantoin, allantoate) for long-distance N transport—especially in legumes—with xanthine dehydrogenase and related enzymes steering flux. During seed germination, stored RNA/DNA are rapidly hydrolyzed; broad-specificity nucleosidases mobilize uridine, xanthosine, and others to supply nucleotide synthesis and ureide production precisely when cell division surges. Under drought or salinity, plants reprogram nucleotide turnover—upshifting salvage and purine catabolism—to balance energy, redox, and osmoprotection while reallocating nitrogen to growing tissues. Together, nucleoside signaling, salvage, and degradation form an integrated network that tunes growth, resilience, and developmental timing—offering targets (transporters, nucleosidases, ureide pathway enzymes) for crop improvement in yield stability and stress tolerance.
Daily Life with Nucleosides: Supplements, Umami, Caffeine, and mRNA Vaccines
From the pantry to the clinic, nucleosides touch daily life. Infant formula often adds dietary nucleotides to support gut and immune development; adult “immune” supplements also market yeast-derived nucleotides for recovery. In food science, RNA breakdown yields IMP and GMP—nucleotide flavor enhancers behind umami in meat, mushrooms, and broths. Caffeine, a purine cousin, blocks adenosine receptors to boost alertness. In healthcare, urine nucleoside panels are emerging as non-invasive biomarkers for cancer monitoring, while purine metabolism links high-purine diets to uric acid and gout (managed with xanthine oxidase inhibitors). Biotech uses modified nucleosides—e.g., pseudouridine in mRNA vaccines—for stability and low innate immunity. Together, these examples show nucleosides’ roles in nutrition, flavor, energy, and precision medicine.
How to Measure Nucleosides: LC–MS/MS and Other Analytical Methods
LC–MS/MS is the gold standard for nucleoside quantification, combining chromatographic separation with targeted mass transitions for high sensitivity and specificity. HPLC-UV remains useful for routine or legacy assays, while capillary electrophoresis supports rapid, charge-based separations in targeted workflows. NMR offers structure-level confirmation and reproducible profiling, and immunoassays enable quick screens for select targets. Emerging electrochemical biosensors provide real-time readouts. Together, these methods balance sensitivity, multiplexing, and practicality—most labs lead with LC–MS/MS and complement it with HPLC/CE for orthogonal checks and NMR for structure confirmation.
Table. Comparison of nucleoside detection methods.
|
Method |
Core principle |
Sensitivity (typical) |
Best for |
Key pros |
Key limitations |
|
LC–MS/MS (triple quad, dMRM) |
LC separation + mass transitions (MRM) |
pg–fg/mL |
Quantitative panels in complex biofluids (urine, serum, cell extracts) |
Highest specificity/sensitivity; large panels possible; wide linear range |
Higher cost; requires isotope-labeled internal standards and expertise |
|
HPLC–UV |
LC + UV (~254 nm) |
ng–µg/mL |
Routine/legacy assays; abundant nucleosides; fraction preparation |
Simple, robust, low cost |
Limited sensitivity/selectivity; co-elution risk |
|
CE–LIF |
Charge/size separation + fluorescence derivatization |
ng–pg/mL (with derivatization) |
Purity checks; targeted research assays |
Fast runs; low sample consumption |
Requires derivatization; less suited to complex matrices |
|
NMR (^1H/^13C) |
Nuclear magnetic resonance spectroscopy |
µM–mM |
Structural confirmation; non-destructive profiling |
Highly reproducible; structure-level insight |
Low sensitivity; expensive instrumentation |
|
Immunoassay (ELISA) |
Antibody-based detection |
ng–pg/mL (analyte-dependent) |
Quick screening of specific targets (e.g., pseudouridine) |
Simple workflow; scalable |
Cross-reactivity; narrower dynamic range than MS |
|
Biosensors (electrochemical) |
Biorecognition → electrical signal |
nM–µM (prototype-dependent) |
Real-time adenosine/related readouts; point-of-need |
Rapid, portable; minimal sample volume |
Limited analyte scope; matrix effects, calibration needed |
Nucleosides are small molecules with outsized impact: they encode and regulate life, power frontline antivirals and chemotherapies, and reveal disease as non-invasive biomarkers. In plants, they direct growth and nitrogen flux. With modern LC–MS/MS profiling, these signals become quantifiable pathways, enabling precision insights from oncology to crop science. As analytical tools mature, nucleoside data will increasingly guide diagnosis, therapy selection, and bioengineering—bridging genotype and phenotype to inform real-world decisions.
Explore Nucleoside Profiling with MetwareBio
Ready to turn nucleosides into answers? MetwareBio delivers high-coverage, high-sensitivity LC–MS/MS metabolomics with rapid turnaround, and expert interpretation across serum, urine, plants, and microbes. We also tailor methods for rare or novel modifications. Contact us to design your study, benchmark biomarkers, or validate engineered pathways—and translate metabolomic readouts into clear, reproducible insights that guide better decisions, refine hypotheses, and deepen understanding for patients, crops, and communities.
References
- Ding J, Li H, Liu Y, et al. OXCT1 Enhances Gemcitabine Resistance Through NF-κB Pathway in Pancreatic Ductal Adenocarcinoma. Front Oncol. 2021;11:698302. Published 2021 Nov 5. doi:10.3389/fonc.2021.698302
- Hsu WY, Chen CJ, Huang YC, Tsai FJ, Jeng LB, Lai CC. Urinary nucleosides as biomarkers of breast, colon, lung, and gastric cancer in Taiwanese. PLoS One. 2013;8(12):e81701. Published 2013 Dec 19. doi:10.1371/journal.pone.0081701
Read more
- Malic Acid vs. Citric Acid: The Powerhouse Acids in Your Favorite Fruits
- Fumaric Acid Unveiled: From Nature's Palette to Therapeutic Potential
- Pyruvic Acid: A Key Player in Cellular Metabolism and Health
- Lactic Acid: Key Roles in Human Metabolism, Diseases, and Health Implications
- Cholic Acid: The Essential Bile Acid Impacting Digestion and Health
- Kynurenine: The Hidden Metabolite Linking Immunity, Mental Health, and Disease Prevention
- Understanding Glycine: Its Metabolism and Vital Role in Human Well-Being
- Leucine: The Branched-Chain Amino Acid That Fuels Muscle Growth
- Unveiling Ornithine: Beyond the Urea Cycle, A Multifaceted Player in Health
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


