Peptidomics: Unlocking the Hidden Language of the Proteome for Precision Medicine
While genomics provides the blueprint and proteomics surveys the vast landscape of proteins, a crucial layer of dynamic biological information flows just beneath the surface. This layer consists of a diverse and bioactive universe of endogenous peptides—short chains of amino acids that act as hormones, neurotransmitters, immune signals, and disease indicators. Peptidomics, the dedicated, system-wide study of these native peptides, deciphers this hidden language, offering unparalleled insights into real-time physiological and pathological processes. By capturing the direct products of protein processing and degradation, peptidomics moves beyond static protein inventories to reveal the active, functional state of the proteome. This blog serves as your comprehensive guide to understanding how peptidomics analysis is revolutionizing biomarker discovery and therapeutic development in the era of precision medicine.
1. What is Peptidomics and Why It’s Different
To fully appreciate the power of peptidomics, we must first define its unique territory within the multi-omics landscape. Unlike the broad surveys of its sibling fields, peptidomics zooms in on a specific, functionally vibrant subset of molecules. This chapter will clarify what constitutes the peptidome, explain its cellular origins, and distinguish it from related disciplines, setting the foundation for understanding its applications.
1.1 Definition and Scope of Peptidomics
Peptidomics is defined as the comprehensive, qualitative, and quantitative analysis of all native, low-molecular-weight peptides (typically < 10 kDa) present in a biological system at a given time—collectively known as the peptidome. This includes a fascinating array of molecules with distinct roles:
i. Bioactive Peptides: The functional elite, including neuropeptides (e.g., substance P, beta-endorphin) that mediate neural communication, and hormone peptides (e.g., insulin, glucagon) that regulate systemic physiology.
ii. Signaling and Regulatory Peptides: Such as cytokines and growth factors.
iii. MHC-Associated Peptides: Peptides presented on cell surfaces by Major Histocompatibility Complex (MHC) molecules, crucial for immune surveillance.
iv. Processing Intermediates and Degradation Products: Peptides generated from the precise cleavage of precursor proteins (prohormones) or from general protein turnover.
The key unifying principle is that these peptides are endogenously generated by specific proteolytic processing within the organism, not by artificial digestion in a lab vial. This makes their profile a direct readout of in vivo protease activity and cellular events.
1.2 The Biological Origins of the Peptidome
The endogenous peptidome is not random; it is a meticulously generated signature of cellular and extracellular activity. Its sources include:
- Prohormone Processing: Many peptide hormones and neuropeptides are synthesized as inactive precursors (e.g., pro-opiomelanocortin) and undergo regulated cleavage by prohormone convertases in secretory vesicles.
- Protease-Mediated Cleavage: Extracellular or intracellular proteases (e.g., matrix metalloproteinases, caspases, kallikreins) selectively cleave proteins, releasing peptide fragments that can have new functions or serve as biomarkers of protease activity, such as in cancer or inflammation.
- Protein Degradation: Peptides generated by the proteasome and other degradation machinery, including those presented by MHC class I molecules.
- Secreted Peptides: Active release of peptides from cells for paracrine or endocrine signaling.
- Intracellular Signaling Peptides: Certain peptides derived from non-canonical sources, like mitochondrial or ribosomal proteins, can have intracellular regulatory roles.
Understanding these origins frames the peptidome not as noise, but as a rich source of functional information about health, disease state, and cellular communication.
1.3 Peptidomics vs. Proteomics vs. Metabolomics
It's crucial to differentiate peptidomics from the more familiar fields of proteomics and metabolomics, as this distinction underpins its unique value.
Proteomics focuses on intact proteins—their identity, quantity, modifications, and interactions. A common proteomic workflow involves using an enzyme like trypsin to digest proteins into predictable tryptic peptides for mass spectrometry analysis. Here, peptides are merely analytical proxies for proteins.
Peptidomics, in contrast, studies the endogenous peptides themselves as the primary biological entities of interest. The peptidome sits conceptually and chemically between the proteome and the metabolome. It represents the dynamic interface where precursor proteins are enzymatically processed into active or inactive fragments, such as the conversion of pro-insulin into the active hormone insulin and its by-product, C-peptide.
Metabolomics deals with small molecules (<1.5 kDa) such as sugars, lipids, and amino acids, which are the end-products of metabolism. Many endogenous peptides, however, are bioactive signaling molecules, not mere metabolic intermediates or waste products.
In essence, while proteomics tells you "what proteins are present," peptidomics reveals "what processing events and proteolytic pathways are actively occurring." To clarify how peptidomics analysis differs from proteomics and metabolomics—and when each approach is most informative—Table 1 summarizes the key distinctions across a few core dimensions.
Table 1. Key Differences Between Peptidomics, Proteomics, and Metabolomics
|
Dimension |
Peptidomics |
Proteomics |
Metabolomics |
|
Primary analytes |
Endogenous peptides (bioactive, neuropeptides, hormone peptides) |
Full-length proteins |
Small molecules (metabolites) |
|
Key analytical technique |
LC-MS/MS (shotgun or targeted proteomics) |
LC-MS, GC-MS |
|
|
Focus of study |
Peptide fragments from proteolysis and signaling |
Protein expression, structure, and PTMs |
Metabolic pathways and small molecule profiling |
|
Quantification |
Label-free, isotope standards for peptides |
Label-free, isotope dilution for metabolites |
|
|
Applications |
Peptide biomarker discovery, protease activity, immunopeptidomics |
Mechanism-of-action, protein biomarkers, PTMs |
Disease biomarkers, metabolic mechanism, drug metabolism |
2. Why Peptidomics Matters: Biological Insight and Translational Value
Having defined the unique territory of peptidomics, the critical question arises: what specific insights can this field provide that others cannot? Unlike static molecular inventories, the peptidome functions as a real-time, dynamic reporter of biological activity. This chapter elucidates the core value proposition of peptidomics by detailing how it answers distinct biological questions, bridges molecular signaling with observable phenotypes, and delivers tangible impact in clinical and pharmaceutical settings.
2.1 Capturing Protease Activity and Pathway Dynamics
Traditional proteomics excels at quantifying changes in protein abundance but often obscures a crucial layer of regulation: proteolytic processing. Many proteins are synthesized as inactive precursors or are functionally modulated through specific cleavage events. Peptidomics uniquely accesses this layer by directly measuring the end products of these cuts.
The presence, absence, or relative abundance of specific endogenous peptides serves as a precise digital readout of protease activity in vivo. For instance, the cleavage signature of a matrix metalloproteinase (MMP) in a tumor microenvironment, or the activation profile of caspases during apoptosis, is indelibly recorded in the peptidome. This allows researchers to move beyond asking “Is this protease present?” to answering “Is this protease active, and what are its specific substrates in this pathological context?” This capability makes peptidomics an indispensable tool for studying cancer invasion, inflammatory processes, and neurodegenerative diseases where protease dysregulation is a hallmark.
2.2 A Bridge Between Signaling and Phenotype
Endogenous peptides are primary agents of cellular and systemic communication. A comprehensive peptide profiling approach, therefore, provides a direct window into the active signaling landscape that connects genetic and protein-level changes to functional phenotypes.
By quantifying neuropeptides, researchers can decode altered neuronal communication in psychiatric disorders or pain states. Profiling circulating hormone peptides and adipokines reveals systemic endocrine and metabolic status. Analyzing the secretome and inflammatory peptides (e.g., kinins, complement peptides) from immune cells offers a dynamic view of immune activation or suppression. Unlike measuring mRNA or even parent protein levels, quantifying these bioactive peptides tells us what signals are actually being transmitted. Thus, peptidomics acts as a functional translator, converting complex proteomic data into an understandable narrative of ongoing biological events.
2.3 Clinical and Biopharma Relevance
The translational power of peptidomics is immense, spanning from discovery to development and monitoring.
i. Biomarker Discovery: The peptidome of easily accessible biofluids like plasma, serum, or urine is a treasure trove for peptide biomarker discovery. Disease-specific protease activities generate unique peptide fragments that can serve as highly specific diagnostic, prognostic, or therapeutic monitoring signatures. Their small size and stability (when properly collected) make them excellent candidates for liquid biopsy applications.
ii. Drug Development & Target Validation: Peptidomics is pivotal in the era of peptide-based therapeutics. It can be used to monitor the in vivo processing and stability of therapeutic peptides, identify novel bioactive peptide drug candidates from natural sources, and validate the downstream effects of protease inhibitor drugs by assessing changes in their native substrate cleavage patterns.
iii. Pharmacodynamics/Toxicity Monitoring: Shifts in the peptidome can serve as sensitive early indicators of drug efficacy or mechanism-based toxicity, reflecting pathway modulation before gross phenotypic changes occur.
iv. Quality Attribute Monitoring: In biopharmaceutical manufacturing, peptidomics can monitor unwanted proteolytic degradation of protein therapeutics (e.g., monoclonal antibodies) throughout the production process, ensuring product quality and consistency.
In essence, peptidomics transforms the peptidome from a biological curiosity into a quantifiable, actionable matrix for advancing precision medicine and biopharmaceutical innovation.
3. Peptidomics Experimental Workflow: From Sample to LC–MS/MS Analysis
The translational potential of any peptidomics analysis is only as strong as its foundational workflow. From the moment of sample collection to the final data output, each step presents specific challenges that can compromise data integrity. This chapter outlines the critical phases of a robust peptidomics pipeline, emphasizing the stringent controls necessary to ensure your results are both biologically meaningful and technically reproducible—enabling confident peptide biomarker discovery and mechanistic insight.
3.1. Mastering Pre-Analytical Variables
The greatest risk to a successful study occurs before the sample is ever frozen. The endogenous peptidome is highly dynamic, susceptible to rapid ex vivo degradation by active proteases. Therefore, standardized protocols are non-negotiable. For biofluids like plasma, serum, or CSF, immediate protease inhibition and consistent low-temperature processing are critical. Strict limits on freeze-thaw cycles, the use of low-adsorption collection tubes, and defined centrifugation conditions are equally vital to "freeze" the true biological signal. Variability here is the primary source of irreproducible results, making detailed SOPs for each sample type (tissue, cells, urine) a key marker of a quality-focused service provider.
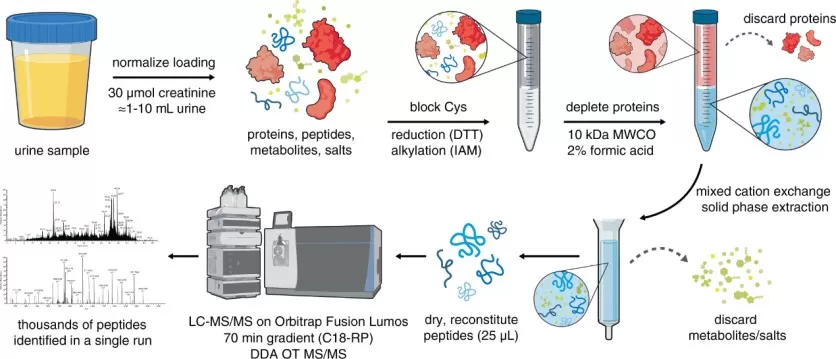
An efficient chemical extraction and LC–MS/MS workflow for urinary peptidomic analysis.
Image reproduced from Palanski et al., 2022, Nature Comunications, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
3.2. Strategic Extraction and Cleanup
Effective analysis requires isolating the native peptide fraction from a complex matrix. The chosen strategy must balance depth of coverage with specificity. Common techniques include protein precipitation for general deproteinization, solid-phase extraction (SPE) for desalting and concentration, or ultrafiltration for size-based separation. For plasma/serum, immunoaffinity depletion of high-abundance proteins can enhance detection of lower-abundance peptides, but it must be applied judiciously to avoid unintentionally removing bound bioactive peptides. The ultimate goal is a clean, MS-compatible sample that faithfully represents the original peptidomic profile.
3.3. Optimized LC-MS/MS Acquisition
The inherent complexity of the peptidome—with its wide length distribution, diverse modifications, and non-tryptic sequences—demands tailored instrument methods. Chromatography typically requires long, shallow gradients on nano-flow systems for sufficient separation. For mass spectrometry, the choice between Data-Dependent Acquisition (DDA) and Data-Independent Acquisition (DIA) is crucial. While DDA is excellent for initial discovery, DIA (e.g., SWATH-MS) is increasingly favored for biomarker discovery due to its superior quantitative consistency and comprehensive data recording across large sample cohorts.
3.4. Rigorous Quantification & QC Framework
Trustworthy quantification is the bridge to biological interpretation. Label-free quantification offers flexibility for large studies, whereas isobaric tagging (e.g., TMT) enables multiplexing but requires advanced instrumentation to mitigate ratio compression. Regardless of the method, a systematic Quality Control (QC) design is essential. This includes running pooled QC samples throughout the sequence to monitor instrument stability, incorporating technical replicates, and applying batch-correction algorithms when needed. This rigorous system suitability monitoring ensures the reported differences are biologically driven, not technically artefactual.
4. Peptidomics Data Analysis and Interpretation: Bioinformatics and Functional Insights
The raw output of a LC-MS/MS peptidomics run is a complex dataset that holds the key to biological discovery. The transition from spectral peaks to mechanistic understanding requires a sophisticated bioinformatics pipeline designed to address the unique challenges of peptide identification and interpretation. This chapter details how expert analysis overcomes these hurdles to transform your data into reliable, actionable biological insights, a critical phase where the true value of your investment is realized.
4.1. Overcoming Peptide Identification Challenges
Unlike standard proteomics, peptidomics faces the “no-enzyme” search problem, where peptides can have any N- and C-terminus, exploding the potential search space and false-positive risk. The key to trustworthy results lies in a stringent analytical pipeline. This involves specialized database searches with tailored parameters, rigorous False Discovery Rate (FDR) control applied specifically to the peptidome context, and advanced de-redundancy algorithms to cluster related peptide sequences and variants. This multi-layered validation ensures that the reported identifications form a solid, credible foundation for all subsequent analysis.
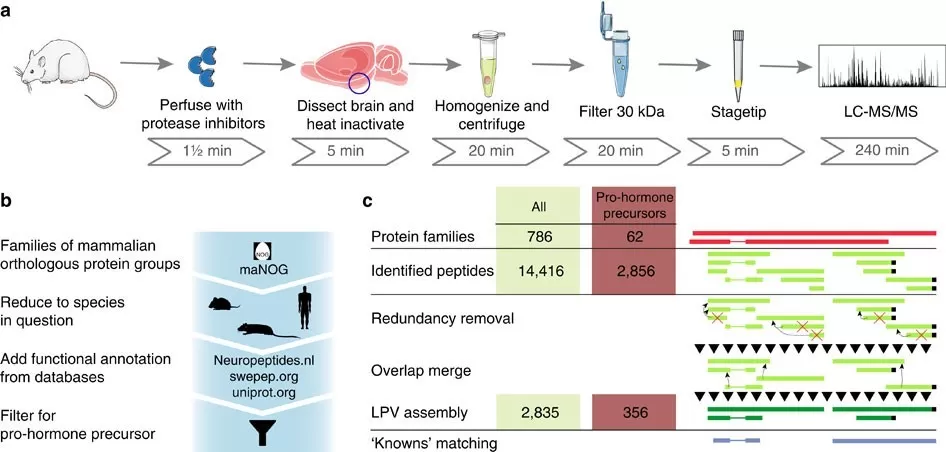
Analytical framework for analysis of endogenous peptides.
Image reproduced from Secher et al., 2016, Nature Comunications, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
4.2. Characterizing PTMs and Peptide Variants
The functional activity of endogenous peptides is often dictated by specific post-translational modifications (PTMs). A proficient peptidomics analysis must therefore accurately detect and localize critical modifications such as C-terminal amidation (activating many neuropeptides), N-terminal acetylation or pyroglutamate formation, and phosphorylation. This capability reveals the concept of peptidoforms—distinct molecular entities derived from a single precursor protein through differential processing and modification. Capturing this detailed landscape is essential for understanding precise bioactive states and disease-associated molecular signatures.
4.3. From Lists to Biology: Functional Interpretation
Moving beyond a simple list of differential peptides requires context-aware bioinformatics. Instead of generic enrichment analysis, meaningful interpretation involves mapping peptides to precursor proteins and then analyzing the specific protein domains or pathways represented by the cleavage events. A powerful application is protease activity inference, where the systematic analysis of peptide termini signatures can predict which proteases were active in the original sample, directly linking peptidomic profiles to regulatory biology. This approach, as demonstrated in recent cancer research (Lopez et al., 2023), transforms peptide data into a functional readout of pathway dynamics and potential therapeutic targets.
5. Frontier Applications and Case Studies of Peptidomics
The theoretical potential of peptidomics is now being realized in laboratories and clinics worldwide, translating complex peptide signatures into actionable insights. This chapter explores the cutting-edge applications where peptidomics is delivering tangible breakthroughs, from redefining disease diagnosis to powering the next generation of personalized therapies. Through concrete case studies, we will see how the analysis of the peptidome is providing unique solutions to longstanding challenges in biomedicine and beyond.
5.1 Disease Biomarker Discovery (Cancer, Neurodegeneration, Cardiovascular Disease)
In clinical peptidomics, LC-MS/MS peptidomics analysis captures the endogenous peptidome—a dynamic readout of protease activity, tissue remodeling, and disease-specific protein degradation—making it highly suited for peptide biomarker discovery when proteins or metabolites are less responsive. A strong example comes from a large-scale wound-infection setting where peptidomics enabled early, mechanism-linked stratification: Hartman and colleagues introduced a peptide-clustering strategy that reduced peptidomics dimensionality by ~95% and markedly improved cross-sample comparability, then applied it to porcine infected wounds and human non-healing wounds. Their analysis revealed phenotype- and pathogen-associated peptide regions detectable at the earliest stages of bacterial colonization, demonstrating how peptide profiling can surface actionable diagnostic signatures from complex clinical biofluids (Hartman et al., 2024).
5.2 Drug Development and Target Identification
In drug R&D, peptidomics-style peptide profiling helps answer two practical questions: which peptide forms actually exist in vivo (stability, degradation, distribution) and which enzymatic processes shape them (target/protease involvement, mechanism-of-action signals). This is especially relevant for peptide therapeutics such as GLP-1 analogs, where subtle structural changes can dramatically alter half-life, tissue exposure, and safety margins. In an open-access study, Oh and colleagues developed a sensitive LC–MS/MS workflow to quantify the therapeutic GLP-1 analog liraglutide in rat plasma and brain regions, enabling pharmacokinetic modeling and tissue distribution assessment. Importantly, they reported low overall brain penetration but region-dependent differences (e.g., higher signal in hypothalamus), illustrating how LC–MS/MS peptide profiling can de-risk CNS hypotheses and refine dosing or formulation strategies during development (Oh et al., 2023).
5.3 Immunopeptidomics and Personalized Medicine (MHC-Bound Peptides)
Immunopeptidomics is a specialized branch of peptidomics that uses LC-MS/MS to directly identify peptides presented by MHC/HLA molecules, providing experimental evidence of what the immune system can “see”—a key advantage over prediction-only pipelines. This makes immunopeptidomics foundational for personalized oncology, including neoantigen vaccine selection, T-cell therapy targeting, and immune monitoring. A recent open-access Science Advances study integrated large immunopeptidomics collections across tumors and normal tissues to build a pan-cancer antigen resource and then used MS-guided discovery to prioritize neoantigen candidates with downstream functional validation, illustrating how immunopeptidomics can move from peptide identification to actionable therapeutic hypotheses for precision immunotherapy (Cai et al., 2025).
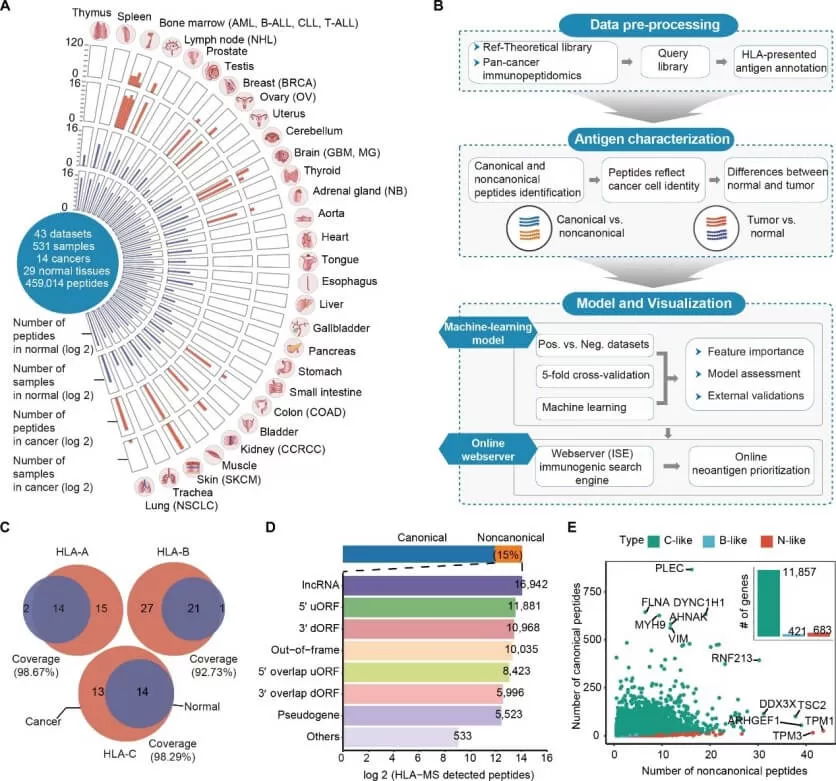
Overview of the HLA-presented tumor-normal immunopeptidome atlas.
Image reproduced from Cai et al., 2025, Science advances
5.4 Food Science and Microbiology (Functional Food Peptides, Antimicrobial Peptides)
Food and microbiology peptidomics focuses on mapping how proteins are enzymatically converted into bioactive peptides, enabling evidence-based development of functional foods, fermentation optimization, and discovery of natural antimicrobial sequences. In practice, LC-MS/MS peptidomics tracks peptide generation across processing timepoints, links peptide motifs to enzymatic specificity, and connects peptide identities to measurable bioactivity (e.g., ACE inhibition, antimicrobial effects). In an open-access Frontiers study, Wang and colleagues applied peptidomics to kefir-grain fermentation of bovine milk, identifying 122 peptides with preferential β-casein hydrolysis hotspots and demonstrating sustained ACE-inhibitory activity even after simulated gastrointestinal digestion (63% activity retained). They also pinpointed multiple Pro-containing ACE-inhibitory peptides, showing how peptidomics can bridge peptide profiling with functional validation for nutraceutical positioning (Wang et al., 2023).
6. Navigating Challenges and Embracing the Future of Peptidomics
Peptidomics has undeniably transitioned from a niche field to a core component of modern multi-omics research, offering unparalleled insights into dynamic biological processes. However, its journey from a powerful discovery tool to a routine clinical and industrial application is paved with both exciting technological frontiers and significant translational hurdles.
6.1 Translational Challenges: The Path to the Clinic
Translating a promising peptide biomarker from laboratory discovery into a rigorously validated, standardized assay for clinical diagnosis or therapeutic monitoring is a long and complex journey. The primary challenges are:
- Standardization and Reproducibility: Peptides, particularly low-abundance bioactive ones, are highly susceptible to pre-analytical variables (e.g., collection time, temperature, protease inhibition). Transitioning from discovery-phase high-resolution LC-MS/MS findings to a stable, clinically viable assay requires establishing extremely stringent standardized operating procedures and quality control systems that perform consistently across different laboratories.
- Clinical Utility Validation: Proving that a peptide biomarker offers diagnostic, prognostic, or predictive value superior to existing clinical standards necessitates meticulously designed, large-scale, multi-center clinical cohort studies. This involves significant cost and complex project management.
- Data Interpretation and Mechanistic Linkage: The key to demonstrating translational value lies in moving beyond a simple list of differential peptides. The critical challenge is robustly linking vast peptidomics datasets to clear biological mechanisms or clinical phenotypes.
6.2 Technological Frontiers: The Next Resolution Revolution
To address these challenges, peptidomics technology is undergoing a profound transformation. Cutting-edge developments aim to deliver more precise, comprehensive, and intelligent solutions:
- Higher-Dimensional Analytical Technology: Traditional liquid chromatography-mass spectrometry (LC-MS/MS) is evolving towards "4D" separation. Adding ion mobility separation as a fourth dimension alongside LC and MS/MS significantly enhances the separation capacity and identification accuracy for complex samples. This is particularly powerful for analyzing isomeric peptides and low-abundance species. It forms the core advantage of platforms like the 4D LC-MS/MS (e.g., timsTOF) with TIMS-PASEF® technology used by MetwareBio, enabling ultra-sensitive detection and more accurate peptide identification.
- Spatial and Single-Cell Resolution: Conventional "bulk" homogenization analysis loses the spatial distribution information of peptides within tissue or cell populations. Emerging fields like spatial peptidomics and single-cell peptidomics aim to reveal the cell-cell communication functions of peptides in specific contexts like the tumor microenvironment or neural circuits, offering the ultimate perspective for understanding their biological function.
- AI and Big-Data Drive: Faced with massive datasets and complex identification needs, Artificial Intelligence (AI) and machine learning are being deployed for peptide spectrum prediction, precise identification, bioactivity prediction, and cross-omics data integration. AI promises to dramatically enhance the depth and efficiency of data analysis, uncovering patterns and associations beyond human discernment.
6.3 Partner with MetwareBio for Your Peptidomics Success
MetwareBio's Peptidomics Service is designed to be your strategic partner in overcoming these translational challenges. Our service is built on a cutting-edge 4D LC-MS/MS platform (Bruker timsTOF HT with TIMS-PASEF® technology and ddaPASEF acquisition), enabling ultra-sensitive detection and accurate quantification of endogenous peptides. We support a broad range of sample types—from tissues and biofluids to cells and plants—with flexible, optimized protocols. Our comprehensive bioinformatics pipeline ensures data transforms into biological insight through functional annotation, pathway enrichment, and network analysis. Backed by extensive experience across biomedical research, microbiology, plant science, and nutrition, our expert team provides end-to-end support to power your biomarker discovery and mechanistic studies.
Reference
- Secher, A., Kelstrup, C. D., Conde-Frieboes, K. W., Pyke, C., Raun, K., Wulff, B. S., & Olsen, J. V. (2016). Analytic framework for peptidomics applied to large-scale neuropeptide identification. Nature communications, 7, 11436. https://doi.org/10.1038/ncomms11436
- Palanski, B. A., Weng, N., Zhang, L., Hilmer, A. J., Fall, L. A., Swaminathan, K., Jabri, B., Sousa, C., Fernandez-Becker, N. Q., Khosla, C., & Elias, J. E. (2022). An efficient urine peptidomics workflow identifies chemically defined dietary gluten peptides from patients with celiac disease. Nature communications, 13(1), 888. https://doi.org/10.1038/s41467-022-28353-1
- Hartman, E., Forsberg, F., Kjellström, S., Petrlova, J., Luo, C., Scott, A., Puthia, M., Malmström, J., & Schmidtchen, A. (2024). Peptide clustering enhances large-scale analyses and reveals proteolytic signatures in mass spectrometry data. Nature communications, 15(1), 7128. https://doi.org/10.1038/s41467-024-51589-y
- Oh, H. S., Choi, M., Lee, T. S., An, Y., Park, E. J., Kim, T. H., Shin, S., & Shin, B. S. (2023). Pharmacokinetics and brain distribution of the therapeutic peptide liraglutide by a novel LC–MS/MS analysis. Journal of Analytical Science and Technology, 14, 19. https://doi.org/10.1186/s40543-023-00382-5
- Cai, Y., Gong, M., Zeng, M., Leng, F., Lv, D., Guo, J., Wang, H., Li, Y., Lin, Q., Jing, J., Zhang, Y., Xu, J., & Li, Y. (2025). Immunopeptidomics-guided discovery and characterization of neoantigens for personalized cancer immunotherapy. Science advances, 11(21), eadv6445. https://doi.org/10.1126/sciadv.adv6445
- Wang, B., Xiao, S., Cai, Y., Chen, X., & Wang, J. (2023). Peptidomics approaches to the discovery and ACE inhibitory effect of casein peptides derived from fermented bovine milk by kefir grains. Frontiers in Sustainable Food Systems, 7, 1208970. https://doi.org/10.3389/fsufs.2023.1208970
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


