Bypassing "Undruggable" Targets: How Phosphoproteomics Illuminates Downstream Pathways for Cancer Therapy
KRAS, MYC, TP53—high-frequency cancer drivers remain notoriously difficult to drug, largely due to structural constraints, adaptive rewiring, and the absence of obvious binding pockets. A practical route forward is to shift the therapeutic focus from the driver itself to the signaling circuitry it controls. Phosphoproteomics offers a direct way to map this circuitry by measuring phosphorylation events—the functional readout of pathway activity—at scale. As precision oncology matures, it is increasingly clear that driver mutations alone provide an incomplete picture: sequencing indicates what could happen, whereas phosphoproteomics reveals what is happening in real time. This distinction helps explain why tumors with the same mutation can show markedly different drug responses, and why resistance may emerge without new genetic alterations. By tracking phosphorylation “traffic,” phosphoproteomics identifies active hubs, bottlenecks, and bypass routes—often exposing druggable kinase nodes downstream of undruggable drivers [1][2].
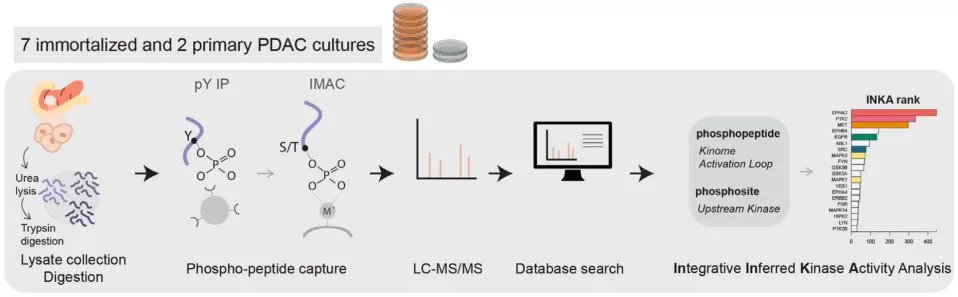
Phosphoproteomic workflow of phosphopeptide enrichment and data processing
Image reproduced from Vallés-Martí et al., 2023, Cell Reports, licensed under the terms of the Creative Commons CC-BY license.
Why Phosphorylation Matters in Driver-Mutation Biology
Phosphorylation functions as a universal control language for cellular signaling, making it essential for understanding how driver mutations reshape oncogenic states. Many canonical drivers (e.g., KRAS, EGFR) exert their effects by persistently engaging pathways such as MAPK or PI3K, but the most faithful way to quantify that engagement is through phosphorylation dynamics rather than DNA changes alone. A mutation may be present, yet pathway flux can be muted by feedback inhibition, lineage context, or compensatory signaling. Conversely, strong pathway activation can occur without a clear driver mutation due to kinase amplification, altered phosphatase activity, or microenvironmental cues. Phosphoproteomics captures these functional realities by measuring pathway output directly, thereby distinguishing genetic potential from biological execution [2].
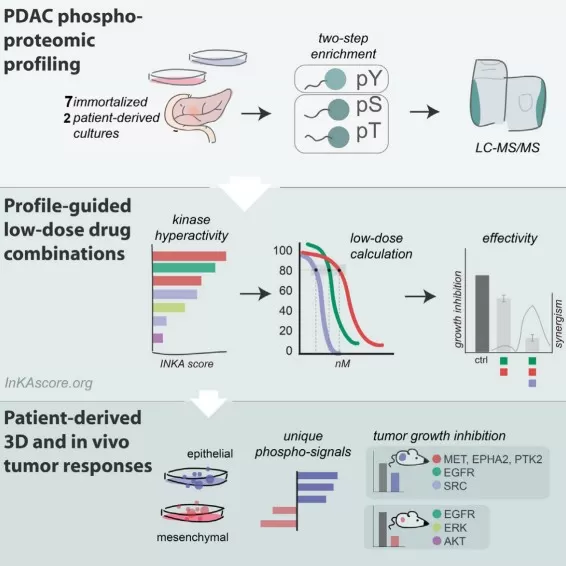
Phosphoproteomics guides effective low-dose drug combinations
Image reproduced from Vallés-Martí et al., 2023, Cell Reports, licensed under the terms of the Creative Commons CC-BY license.
Importantly, relying on sequencing alone can obscure actionable intervention points and resistance mechanisms. Phosphoproteomics can identify which kinases or receptor nodes are truly active, enabling rational targeting of downstream “druggable” components even when the initiating driver is pharmacologically intractable. In EGFR-driven settings, for example, resistance may arise through activation of alternative kinase programs that are not obvious from genomics. By pinpointing these active hubs, phosphoproteomics supports combination strategies—pairing a primary targeted agent with inhibitors against emergent bypass kinases—to reduce escape and improve durability [3][4].
In practice, this paradigm can reveal dependencies missed by genomics. In pancreatic ductal adenocarcinoma, phosphoproteomic profiling has been used to prioritize signaling vulnerabilities and guide effective drug combinations, illustrating how functional pathway readouts can translate into actionable treatment hypotheses.
What Is Cancer Phosphoproteomics, and What Can It Answer?
Cancer phosphoproteomics is a high-throughput LC–MS/MS–based approach that quantifies phosphorylation sites across thousands of proteins, enabling system-level inference of signaling activity. Typical workflows enrich phosphopeptides (e.g., IMAC or TiO₂), quantify phosphosites across conditions, and interpret changes through kinase-substrate relationships and network models. Because phosphorylation is proximal to signaling control, phosphoproteomics provides functional information that standard genomics—and even total proteomics—often cannot fully recover [2].
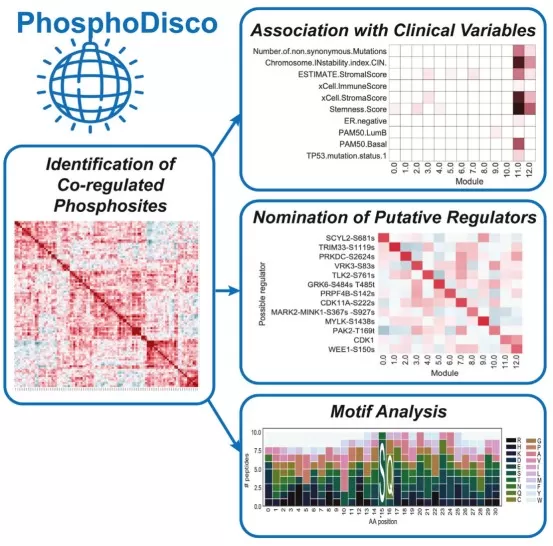
PhosphoDisco Toolkit for Co-regulated Phosphorylation Module Discovery
Image reproduced from Schraink et al., 2023, Mol Cell Proteomics, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
In precision oncology, phosphoproteomics is particularly useful for addressing the following questions:
|
Question |
What phosphoproteomics reveals |
Why it matters clinically |
|
Which pathway is functionally activated in this tumor? |
Direct evidence of pathway output (e.g., MAPK vs PI3K activity) from phosphosite shifts |
Confirms whether a mutation is translating into actionable signaling |
|
Which kinase(s) act as dominant hubs? |
Kinase-activity inference via substrate enrichment/motif patterns |
Prioritizes druggable kinases as intervention points |
|
How does signaling reroute under therapy? |
Adaptive bypass activation, feedback loops, RTK switching |
Guides combination regimens to block escape routes |
|
Can we build functional biomarkers? |
Phosphosite signatures associated with response/prognosis |
Enables stratification beyond mutation status |
These capabilities are illustrated across multiple disease contexts. In HER2-negative breast cancer, phosphoproteomics has been used to identify signaling programs associated with therapeutic sensitivity that are not obvious from genomics alone [3]. In T-cell acute lymphoblastic leukemia (T-ALL), phosphoproteomic profiling revealed targetable kinase dependencies and informed combination strategies [4]. In KRAS-mutant cancers, phosphoproteomics has been used to nominate combination therapies by revealing pathway co-dependencies and compensatory signaling [5]. Methodologically, causal and multiomics-aware analysis frameworks further strengthen interpretability by linking phosphosite changes to upstream regulators and clinical phenotypes [6] [7].
From Samples to Druggable Clues—A Practical Workflow
A typical phosphoproteomics pipeline converts patient samples or model systems into therapeutic hypotheses through an end-to-end, testable process:
Clinical sample / model → phosphopeptide enrichment → LC–MS/MS → computational inference → experimental validation
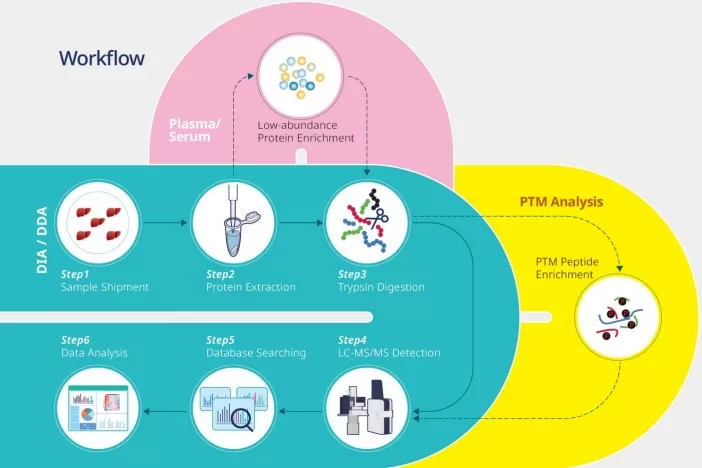
A practical workflow for phosphoproteomics analysis
Step 1: Study design and sample handling
Rigorous design is essential because phosphorylation is transient and sensitive to pre-analytical variables. Biological replicates, batch randomization, and consistent processing reduce false discoveries and improve reproducibility. High-quality handling (rapid freezing, standardized lysis conditions, phosphatase inhibition) preserves true signaling states. Well-controlled tumor–normal comparisons have enabled identification of patient-specific kinase dependencies, supporting individualized target nomination in settings such as cholangiocarcinoma [8].
Step 2: Enrichment and mass spectrometry acquisition
After protein digestion, phosphopeptide enrichment (IMAC/TiO₂) increases detection depth by isolating phosphorylated species from complex backgrounds. Modern quantitative acquisition strategies improve cross-sample consistency and coverage, enabling large-scale phosphosite measurement in heterogeneous tumor models. Broad KRAS-mutant phosphoproteomic landscapes have demonstrated how this depth can expose conserved kinase programs that are therapeutically actionable [5].
Step 3: Computational interpretation to actionable targets
Downstream analysis typically includes differential phosphosite testing, kinase-activity inference, and network-based prioritization to identify hubs and co-regulated modules. Toolkits for module discovery can reveal coordinated phosphorylation programs that map to pathway states and drug vulnerabilities [2]. Integrative, causal approaches help connect phosphoproteomic changes to upstream regulators and phenotypes—especially valuable in resistance settings where signaling rewires without new mutations [8]. Prioritized targets are then validated using perturbation assays (inhibitors, CRISPR/siRNA), organoids, or in vivo models.
High-Value Use Cases for Target Discovery and Therapy Design
Phosphoproteomics is most impactful where genomics underspecifies mechanism and actionability. Below are five high-yield scenarios supported by landmark case studies.
Scenario 1: Targeting druggable kinases downstream of undruggable drivers
Driver mutations like KRAS or RET are notoriously hard to drug directly, but their downstream kinases (e.g., MAPK components) often have existing inhibitors. Phosphoproteomics maps these activations, revealing "druggable" nodes where therapies can be redirected. A seminal study in T-cell acute lymphoblastic leukemia (T-ALL) used global phosphoproteomics to profile cell lines, identifying SRC-family kinases as dominant hubs despite the absence of actionable mutations. This led to testing SRC inhibitors, which showed efficacy in preclinical models, demonstrating how functional signaling maps can convert “undruggable” genetics into druggable intervention points [4]. This approach is vital because mutations alone don't guarantee pathway activity—phosphoproteomics provides functional proof, reducing trial failure rates.
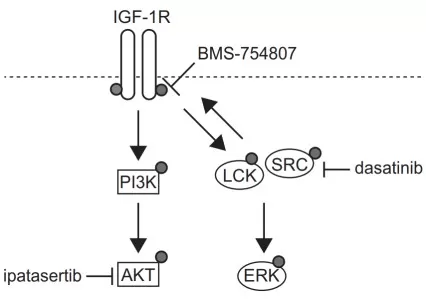
Potential targeting of the INSR/IGF-1R axis and the SFKs signaling
Image reproduced from Cordo et al., 2022, Nature Commun, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
Scenario 2: Identifying bypass pathways to design rational combinations
Single-target therapies often fail due to adaptive "rewiring," where inhibition of one node activates compensatory pathways. Phosphoproteomics rapidly detects these shifts, suggesting combination strategies. For example, in KRAS-mutant cancers, phosphoproteomic profiling uncovered that MEK inhibition triggers feedback activation of PI3K-AKT signaling. Co-targeting both pathways with inhibitors like trametinib and ipatasertib enhanced efficacy in models, demonstrating how phospho-data prevents escape routes [5]. This scenario is crucial for overcoming intrinsic resistance, as genomics misses such dynamic changes, which occur without new mutations.
Scenario 3: Decoding resistance as network reprogramming rather than single mutations
Resistance frequently emerges through system-level signaling adaptation. In ALK-driven neuroblastoma models, phosphoproteomics supported the identification of vulnerabilities that enabled effective suppression via ATR inhibition, highlighting how functional profiling can uncover non-obvious resistance liabilities [7].
Scenario 4: Developing functional biomarkers for stratification beyond genotype
Beyond mutations, phosphorylation signatures predict drug response and prognosis, enabling precision treatment. In HER2-negative breast cancer, phosphoproteomics stratified patients based on CDK or MAPK pathway activity, identifying subsets responsive to kinase inhibitors, even when genomics was inconclusive. This functional biomarker approach improved survival predictions in trials, emphasizing that "pathway activity" trumps mutation status for stratification. Such signatures are more translatable, as they reflect real-time biology, reducing reliance on inconsistent genomic markers [3][8].
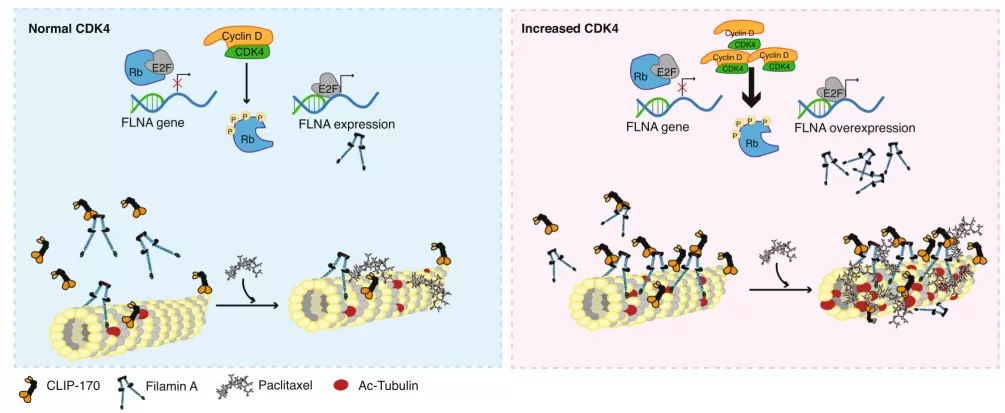
Schematic representing the proposed mechanism of CDK4
Image reproduced from Mouron et al., 2022, Nat Commun, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
Scenario 5: Linking signaling to metabolic remodeling during progression and resistance
Oncogenic signaling can rewire metabolism through phosphorylation-mediated control of metabolic enzymes and stress-response programs. In hepatocellular carcinoma, mechanisms associated with sorafenib resistance have been connected to broader adaptive states, reinforcing the need to interpret resistance through functional pathway remodeling rather than genetics alone [9].
Collectively, these applications show how phosphoproteomics turns “undruggability” from a dead end into a navigation problem: once downstream signaling traffic is measured, therapeutic detours become evidence-based rather than speculative.
References
1. Vallés-Martí A, Mantini G, Manoukian P, Waasdorp C, Sarasqueta AF, de Goeij-de Haas RR, Henneman AA, Piersma SR, Pham TV, Knol JC, Giovannetti E, Bijlsma MF, Jiménez CR. Phosphoproteomics guides effective low-dose drug combinations against pancreatic ductal adenocarcinoma. Cell Rep. 2023 Jun 27;42(6):112581. doi: 10.1016/j.celrep.2023.112581.
2. Schraink T, Blumenberg L, Hussey G, George S, Miller B, Mathew N, González-Robles TJ, Sviderskiy V, Papagiannakopoulos T, Possemato R, Fenyö D, Ruggles KV. PhosphoDisco: A Toolkit for Co-regulated Phosphorylation Module Discovery in Phosphoproteomic Data. Mol Cell Proteomics. 2023 Aug;22(8):100596. doi: 10.1016/j.mcpro.2023.100596.
3. Mouron S, Bueno MJ, Lluch A, Manso L, Calvo I, Cortes J, Garcia-Saenz JA, Gil-Gil M, Martinez-Janez N, Apala JV, et al. Phosphoproteomic analysis of neoadjuvant breast cancer suggests that increased sensitivity to paclitaxel is driven by CDK4 and filamin A. Nat Commun. 2022 Dec 7;13(1):7529. doi: 10.1038/s41467-022-35065-z.
4. Cordo' V, Meijer MT, Hagelaar R, de Goeij-de Haas RR, Poort VM, Henneman AA, Piersma SR, Pham TV, Oshima K, Ferrando AA, Zaman GJR, Jimenez CR, Meijerink JPP. Phosphoproteomic profiling of T cell acute lymphoblastic leukemia reveals targetable kinases and combination treatment strategies. Nat Commun. 2022 Feb 25;13(1):1048. doi: 10.1038/s41467-022-28682-1.
5. Liu Z, Liu Y, Qian L, Jiang S, Gai X, Ye S, Chen Y, Wang X, Zhai L, Xu J, Pu C, Li J, He F, Huang M, Tan M. A proteomic and phosphoproteomic landscape of KRAS mutant cancers identifies combination therapies. Mol Cell. 2021 Oct 7;81(19):4076-4090.e8. doi: 10.1016/j.molcel.2021.07.021.
6. Khorsandi SE, Dokal AD, Rajeeve V, Britton DJ, Illingworth MS, Heaton N, Cutillas PR. Computational Analysis of Cholangiocarcinoma Phosphoproteomes Identifies Patient-Specific Drug Targets. Cancer Res. 2021 Nov 15;81(22):5765-5776. doi: 10.1158/0008-5472.CAN-21-0955. Epub 2021 Sep 22.
7. Szydzik J, Lind DE, Arefin B, Kurhe Y, Umapathy G, Siaw JT, Claeys A, Gabre JL, Van den Eynden J, Hallberg B, Palmer RH. ATR inhibition enables complete tumour regression in ALK-driven NB mouse models. Nat Commun. 2021 Nov 24;12(1):6813. doi: 10.1038/s41467-021-27057-2.
8. Dong Q, Tan M, Zhou Y, Zhang Y, Li J. Causal Inference and Annotation of Phosphoproteomics Data in Multiomics Cancer Studies. Mol Cell Proteomics. 2025 Mar;24(3):100905. doi: 10.1016/j.mcpro.2025.100905.
9. Rodrigo MAM, Michalkova H, Jimenez AMJ, Petrlak F, Do T, Sivak L, Haddad Y, Kubickova P, de Los Rios V, Casal JI, Serrano-Macia M, Delgado TC, Boix L, Bruix J, Martinez Chantar ML, Adam V, Heger Z. Metallothionein-3 is a multifunctional driver that modulates the development of sorafenib-resistant phenotype in hepatocellular carcinoma cells. Biomark Res. 2024 Apr 9;12(1):38. doi: 10.1186/s40364-024-00584-y.
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


