Phosphoproteomics in Drug Discovery: Revealing Hidden Pathways for Targeted Therapies
Introduction: Why Phosphoproteomics Matters in Drug Discovery
Phosphorylation acts as one of the most critical regulatory switches in biology, controlling processes from cell growth and apoptosis to signal transduction in health and disease. When these phosphorylation events are altered, they often drive cancer progression, drug resistance, and other complex disorders. Phosphoproteomics, the large-scale study of phosphorylation across the proteome, provides an unparalleled lens to capture these dynamic modifications. Unlike static genomics or transcriptomics, phosphoproteomics reveals the real-time activity of signaling pathways and drug targets, making it indispensable for modern drug discovery. By mapping phosphorylation landscapes, researchers can identify novel biomarkers, evaluate kinase inhibitor efficacy, and uncover compensatory mechanisms that lead to therapeutic resistance—directly linking molecular changes to clinical outcomes.
The global proteomics market is projected to reach $49 billion by 2028, highlighting its growing role in healthcare innovation. For pharmaceutical companies, biotech researchers, and clinicians, phosphoproteomics offers a roadmap to more effective therapies, fewer side effects, and improved patient outcomes. In this guide, we’ll explore the science, technologies, clinical applications, and emerging trends in phosphoproteomics — with insights into how MetwareBio’s proteomics services can support your research.
The Science of Phosphoproteomics: Decoding Cellular Signals
Phosphorylation is one of the most common and dynamic post-translational modifications, acting as a molecular switch that regulates nearly every aspect of cellular activity—from metabolism and growth to stress responses and communication between cells. By adding or removing phosphate groups, kinases and phosphatases fine-tune signaling pathways such as MAPK, PI3K/AKT, and JAK/STAT, many of which are directly implicated in cancer, immune regulation, and drug resistance.
Phosphoproteomics provides a systematic way to capture and quantify these events on a global scale. Using high-resolution LC–MS/MS on the Bruker timsTOF HT with PASEF technology, researchers can identify thousands of phosphorylation sites in a single experiment, including those present at low abundance. This level of detail makes it possible to see not just which proteins are present, but how they are regulated in real time.
Accurate analysis also depends on advanced computational tools. Site-localization algorithms such as Ascore and phosphoRS, combined with quantification platforms like MaxQuant and Perseus, ensure that phosphorylation sites are assigned with high confidence. This enables scientists to move beyond protein identification and toward decoding the dynamic signaling networks that underlie health and disease.
By mapping these phosphorylation-driven networks, phosphoproteomics provides critical insights into how cells respond to drugs, adapt to therapy, or develop resistance—making it an essential tool in modern biomedical research and drug discovery.
Core Technologies in Phosphoproteomics
Phosphoproteomics relies on a combination of advanced analytical strategies that enable researchers to capture and interpret dynamic phosphorylation events across the proteome. The workflow typically begins with enrichment methods such as immobilized metal affinity chromatography (IMAC) or titanium dioxide (TiO₂), which selectively isolate phosphorylated peptides from complex biological samples. This step ensures that low-abundance phosphorylation sites can be detected with confidence.
Once enriched, samples are analyzed by high-resolution mass spectrometry (LC–MS/MS), which provides sensitive and accurate detection of thousands of phosphorylation sites within a single experiment. Depending on research goals, different acquisition strategies can be applied: broad, discovery-driven approaches for large-scale mapping of signaling networks, and targeted approaches for precise quantification of selected phosphorylation sites.
Finally, bioinformatics tools and pathway analysis platforms are integrated to place the phosphoproteomic data into a biological context. These analyses reveal how phosphorylation events regulate cellular signaling, how pathways are rewired in disease states, and where potential intervention points for drug development may lie.
Together, these technologies form the backbone of modern phosphoproteomics, providing a powerful means to connect molecular signaling with therapeutic discovery.
Clinical Applications: From Biomarkers to Targeted Therapies
Phosphoproteomics serves as a bridge between molecular biology and patient care by linking dynamic signaling events with therapeutic outcomes. Through systematic mapping of phosphorylation, researchers can stratify patients, monitor therapeutic responses, and identify new drug targets. Unlike static genomic data, phosphorylation profiles provide a real-time snapshot of pathway activity, offering unique insights into treatment design.
Biomarker Discovery
Phosphoproteomics provides high-resolution molecular fingerprints that can stratify patients and guide precision diagnostics. For example, an integrated proteogenomic and phosphoproteomic analysis of colorectal cancer identified subtype-specific phosphorylation patterns that correlate with prognosis and therapeutic vulnerabilities (Cell, 2019; PMID: 31031003). More recently, a comprehensive phosphoproteomic atlas of colorectal cancer progression highlighted signaling alterations across disease stages, pointing to candidate biomarkers for patient risk stratification (Cell Reports Medicine, 2024; PMID: 38861363). These studies demonstrate the power of phosphoproteomics in discovering actionable biomarkers for early detection and treatment monitoring.
Kinase Inhibitor Development
Resistance to kinase inhibitors often arises from compensatory pathway rewiring. Phosphoproteomic profiling enables researchers to uncover these adaptive mechanisms. In clear cell renal cell carcinoma, multi-omics and phosphoproteomic analyses revealed distinct molecular subtypes and signaling differences that influence sunitinib response (Nature Communications, 2023; PMID: 37460463). Complementary work using functional phosphoproteomic readouts identified candidate biomarkers predictive of sunitinib benefit (Clinical Proteomics, 2023; PMID: 37940875). By mapping phosphorylation-driven signaling changes, phosphoproteomics accelerates the design of next-generation inhibitors and combination therapies.
Patient Outcomes
Integrating phosphoproteomics into clinical workflows is beginning to shape personalized treatment strategies. A recent prospective study demonstrated that combining reverse-phase protein array (RPPA)-based phosphoproteomics with genomics in molecular tumor boards provided additional or alternative therapeutic options for more than half of enrolled patients (npj Precision Oncology, 2025; PMID: 40234655, PMCID: PMC12000509). This work illustrates how phosphoproteomic signatures can be used in real-world decision-making, supporting more precise therapy selection and improving patient care pathways.
Together, these applications highlight how phosphoproteomics transforms from a discovery tool into a translational engine—providing actionable insights that accelerate drug development and deliver tangible benefits for patients.
Emerging Trends: AI and Single-Cell Phosphoproteomics
The future of phosphoproteomics lies in combining technological innovation with computational intelligence. Traditional workflows have enabled deep coverage of phosphorylation events, but new advances are pushing the boundaries of scale, resolution, and clinical utility.
AI-Driven Proteomics
Phosphoproteomic datasets are vast and complex, often containing tens of thousands of phosphorylation sites. AI and machine learning algorithms now play a critical role in improving peptide identification, site localization, and functional annotation. For example, deep learning–based rescoring tools have been shown to increase phosphosite identification rates by more than 20% compared with conventional approaches (Nature Methods, 2021; PMID: 34140708). Beyond identification, predictive modeling is being used to infer kinase activity, prioritize druggable targets, and simulate network dynamics, accelerating the translation of phosphoproteomic insights into therapeutic strategies.
Single-Cell Phosphoproteomics
Bulk phosphoproteomics provides averaged readouts, but tumor heterogeneity and immune cell diversity demand single-cell resolution. Emerging single-cell phosphoproteomics technologies—powered by highly sensitive mass spectrometry platforms such as Bruker timsTOF HT with PASEF—are beginning to resolve phosphorylation landscapes at the individual cell level. Recent studies have demonstrated that single-cell phosphoproteomic profiling can uncover intratumoral signaling diversity and reveal drug-resistant subpopulations that are invisible in bulk analyses (Nature Biotechnology, 2022; PMID: 35241815). This granular perspective enables precision targeting of signaling pathways that drive therapy resistance.
Together, AI-driven data analysis and single-cell phosphoproteomics mark a paradigm shift, transforming phosphoproteomics from a discovery tool into a high-resolution, predictive platform for drug development and personalized medicine.
Practical Workflow for Phosphoproteomics in Drug Discovery
A successful phosphoproteomics project requires a carefully designed workflow that balances sample preparation, analytical sensitivity, and data interpretation. The process begins with sample preparation and phosphopeptide enrichment, typically using titanium dioxide (TiO₂), immobilized metal affinity chromatography (IMAC), or Fe³⁺-NTA approaches to selectively capture phosphorylated peptides from complex lysates. These enrichment strategies are critical for reducing sample complexity and enhancing the detection of low-abundance phosphorylation events.
Next, enriched samples are analyzed using high-resolution mass spectrometry. At MetwareBio’s U.S. facility, we employ the Bruker timsTOF HT equipped with PASEF technology, which delivers ultrafast acquisition, deeper coverage of phosphoproteomes, and superior sensitivity for low-abundance modifications compared with older Orbitrap-based platforms. This ensures high-confidence identification of thousands of phosphorylation sites in a single run.
Following acquisition, data processing and site localization are performed using validated algorithms such as MaxQuant, Perseus, and Spectronaut, combined with site-localization scoring tools (e.g., Ascore, ptmRS). These tools assign probabilities to phosphorylation sites, ensuring accuracy and minimizing false positives.
Finally, bioinformatics integration links phosphoproteomic data with biological pathways and drug targets. Network analysis platforms (e.g., KEGG, Reactome, PhosphoSitePlus) allow researchers to uncover signaling modules altered in disease or drug treatment. By combining rigorous enrichment methods, advanced instrumentation, and robust computational pipelines, phosphoproteomics provides a practical, end-to-end workflow for accelerating biomarker discovery and targeted therapy development.
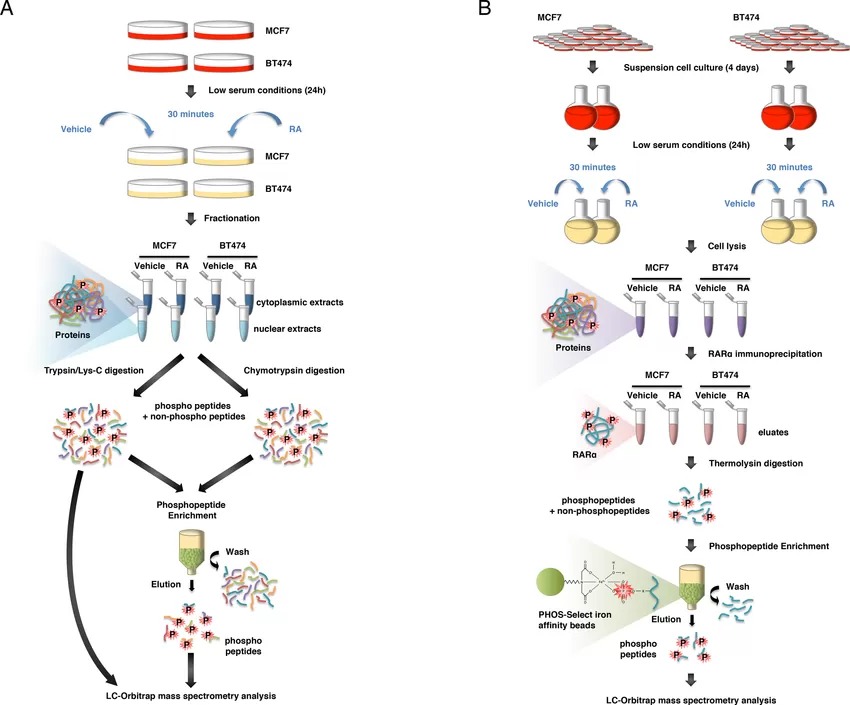
Workflow for the phosphoproteomics strategy.(Carrier et al,. 2016)
Why Choose MetwareBio for Phosphoproteomics Services?
At MetwareBio, we understand that phosphoproteomics requires more than advanced instrumentation—it demands a reliable partner who can deliver data quality, biological insight, and project support from start to finish. Our services are designed to meet the needs of researchers in academia, biotechnology, and pharmaceutical development by combining cutting-edge technology with deep scientific expertise.
-
State-of-the-Art Platform: Our workflows are powered by the Bruker timsTOF HT with PASEF technology, enabling rapid acquisition, high sensitivity, and broad phosphoproteome coverage. This ensures reliable detection of both abundant and low-abundance phosphorylation events.
-
Rigorous Data Processing: All datasets undergo strict quality control, site-localization scoring, and pathway enrichment analysis using widely recognized software tools and databases, ensuring confidence and reproducibility in results.
-
Collaborative Expertise: Our team brings extensive experience in proteomics and bioinformatics, working closely with clients to tailor experimental designs and data interpretation strategies that align with specific research goals.
By choosing MetwareBio, you gain more than a service provider—you gain a scientific partner committed to delivering accurate, reproducible, and biologically meaningful phosphoproteomic insights that can accelerate your discoveries.
Frequently Asked Questions
Q1: What is phosphoproteomics in drug discovery?
Phosphoproteomics is the large-scale study of protein phosphorylation. In drug discovery, it maps cellular signaling pathways to identify druggable targets, enabling precision medicine strategies in cancer, neurology, and beyond.
Q2: How does TMT labeling improve proteomic analysis?
TMT labeling allows multiplexing of up to 18 samples, increasing throughput and improving quantification accuracy. This reduces variability and provides consistent comparative analysis across patient samples or experimental conditions.
Q3: Can AI enhance phosphoproteomics data analysis?
Yes. AI tools such as deep learning models improve peptide site identification, reduce false discovery rates, and integrate proteomics with genomics and transcriptomics for comprehensive insights.
Q4: Why is phosphoproteomics critical for cancer therapies?
Cancer is driven by dysregulated signaling pathways. Phosphoproteomics enables researchers to map phosphorylation-driven kinase networks, guiding the development of targeted therapies that improve patient outcomes.
Q5: How to start a phosphoproteomics project?
Begin by defining your research question and selecting a suitable workflow (e.g., TMT labeling for quantification, DIA for discovery). Partnering with service providers like MetwareBio ensures access to cutting-edge platforms, expert data analysis, and validated protocols.
References
1. Carrier, Marilyn & Joint, Mathilde & Lutzing, Régis & Page, Adeline & Rochette-Egly, Cécile. (2016). Phosphoproteome and Transcriptome of RA-Responsive and RA-Resistant Breast Cancer Cell Lines. PLOS ONE. 11. e0157290. 10.1371/journal.pone.0157290.
2. Aslam B, Basit M, Nisar MA, et al. Proteomics: Technologies and Their Applications. J Chromatogr Sci. 2017;55(2):182–196. doi:10.1093/chromsci/bmw167
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


