What Is Shotgun Proteomics? Principles, Workflow, and Proteomic Data Analysis
Proteomics aims to comprehensively characterize the protein composition of biological systems and understand how proteins change across conditions, time points, or disease states. Among the various proteomic strategies developed over the past decades, shotgun proteomics has emerged as the most widely used and versatile approach for large-scale protein identification and quantification.
Often described as the core implementation of bottom-up proteomics, shotgun proteomics combines enzymatic digestion, high-resolution LC-MS/MS proteomics, and advanced computational analysis to enable global proteome profiling. This article provides a systematic introduction to shotgun proteomics, explaining its principles, experimental workflow, data analysis strategies, and how it relates to other proteomic approaches.
What Is Shotgun Proteomics?
Shotgun proteomics is a peptide-centric proteomic strategy in which complex protein mixtures are enzymatically digested into peptides prior to mass spectrometry analysis. Instead of analyzing intact proteins directly, the method relies on identifying peptides and inferring the corresponding proteins computationally.
The term “shotgun” reflects the untargeted nature of the approach. Rather than focusing on predefined proteins, shotgun proteomic analysis aims to sample and identify as many peptides as possible from a complex biological sample in a single experiment. This makes it particularly well suited for discovery-driven studies, where the goal is to obtain a broad overview of protein expression.
In practice, shotgun proteomics represents the most common form of bottom-up proteomics, distinguishing itself from targeted proteomics and top-down proteomics, which analyze intact proteins without prior digestion. (learn more at: Top-Down vs. Bottom-Up Proteomics)
Shotgun Proteomics as a Core Bottom-Up Proteomics Strategy
To understand the role of shotgun proteomics, it is important to place it within the broader landscape of proteomic methodologies. Bottom-up proteomics refers to any approach in which proteins are first digested into peptides before mass spectrometric analysis. Shotgun proteomics is the dominant bottom-up strategy because it enables high-throughput, unbiased protein identification across diverse sample types.
In contrast, targeted proteomics focuses on the quantitative measurement of predefined peptides or proteins, while top-down proteomics analyzes intact proteins to preserve proteoform-level information. Although each approach has unique advantages, shotgun proteomics remains the preferred choice for global proteome profiling due to its scalability, sensitivity, and compatibility with modern mass spectrometry platforms.
By leveraging peptide-level information, shotgun proteomics achieves deep proteome coverage, making it a foundational method in systems biology, functional genomics, and translational research.
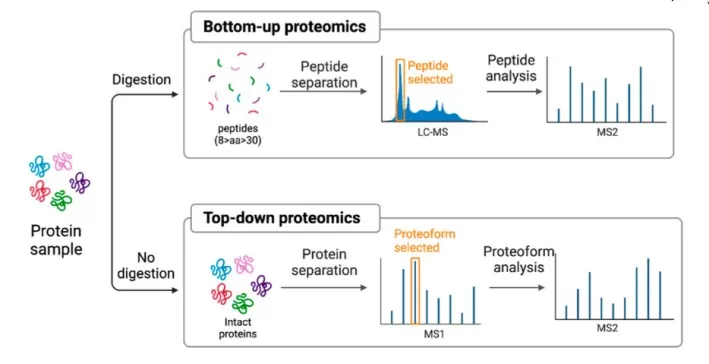
MS-based proteomics is largely divided into bottom-up and top-down approaches.
Image reproduced from Yim and Nestler, 2023, Biomolecules, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
Experimental Workflow of Shotgun Proteomics
The shotgun proteomics workflow consists of several interconnected steps, each of which influences data quality and biological interpretability. While specific protocols may vary, the overall structure is highly standardized across laboratories.
Step One: Protein Extraction and Sample Preparation
Protein extraction represents a critical determinant of shotgun proteomics performance, particularly when working with complex or heterogeneous samples such as tissues, biofluids, or primary cells. The primary goal is to achieve broad and unbiased solubilization of proteins while maintaining compatibility with downstream enzymatic digestion and LC-MS/MS analysis. Following extraction, proteins are typically reduced and alkylated to disrupt disulfide bonds and improve digestion efficiency.
In practice, lysis buffer composition must be carefully optimized. Strong detergents or chaotropic agents can improve protein solubilization, especially for membrane-associated proteins, but may interfere with protease activity or mass spectrometric detection if not adequately removed. Consequently, many shotgun proteomics workflows incorporate cleanup or buffer-exchange steps prior to digestion to minimize chemical interference. Equally important is the control of sample-to-sample variability during preparation. Differences introduced at this stage often propagate through the entire workflow and cannot be corrected computationally. For quantitative shotgun proteomics, standardized protocols and quality control measures at the protein extraction stage are therefore essential.
Step Two: Enzymatic Digestion and Peptide Generation
Enzymatic digestion defines the peptide space that will ultimately be interrogated by mass spectrometry. Trypsin remains the most widely used protease in shotgun proteomics because it generates peptides with predictable charge states and fragmentation behavior, facilitating reliable peptide identification.
However, digestion efficiency is influenced by multiple factors, including protein denaturation, enzyme-to-substrate ratio, incubation time, and temperature. Incomplete digestion can reduce proteome coverage and complicate protein inference, while overdigestion may generate overly short peptides with reduced analytical value.
In recent shotgun proteomics studies, multi-enzyme digestion strategies or sequential digestion approaches have been explored to improve coverage of structurally complex or poorly digestible proteins. Although these strategies increase experimental complexity, they highlight the importance of digestion optimization in maximizing the depth of shotgun proteomic analysis.
Step Three: LC-MS/MS Analysis and Data Acquisition
Following digestion, peptides are separated by liquid chromatography and analyzed by tandem mass spectrometry. In classical shotgun proteomics, data-dependent acquisition (DDA) is used to dynamically select precursor ions for fragmentation based on signal intensity. DDA-based shotgun proteomics benefits from high identification specificity but is inherently stochastic, as not all peptides are selected for fragmentation in every run. This sampling behavior can lead to missing values across large cohorts, particularly for low-abundance peptides.
To address these challenges, advances in chromatography, scan speed, and ion mobility separation have significantly improved peptide sampling efficiency. In parallel, data-independent acquisition (DIA) has emerged as an alternative strategy that retains the discovery-oriented nature of shotgun proteomics while improving reproducibility. As a result, modern shotgun proteomic workflows increasingly integrate both DDA and DIA, depending on study objectives and sample complexity. (learn more at: DDA vs. DIA: The Essential Guide to Label-Free Quantitative Proteomics)
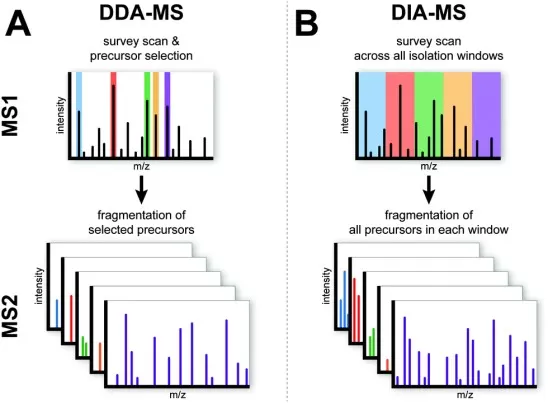
Schematic overview of the DDA-MS and DIA-MS.
Image reproduced from Krasny and Huang, 2021, Molecular omics, licensed under the Creative Commons Attribution 3.0 International License (CC BY 3.0).
Shotgun Proteomics Data Analysis
The transformation of raw LC-MS/MS data into biologically interpretable results is one of the most technically demanding aspects of shotgun proteomics. Data analysis pipelines must balance sensitivity, specificity, and computational robustness to ensure reliable conclusions.
Peptide Identification and Database Searching
Fragmentation spectra acquired during LC-MS/MS analysis are matched against theoretical spectra derived from protein sequence databases. This process, known as peptide identification, is highly sensitive to mass accuracy, fragmentation quality, and database completeness.
To control false identifications, false discovery rate (FDR) estimation is routinely applied using target–decoy strategies. Importantly, FDR control at the peptide level does not automatically translate to protein-level confidence, making careful interpretation essential in shotgun proteomic analysis. Recent developments in spectral library construction and machine learning–based scoring algorithms have substantially improved peptide identification rates, particularly in complex samples. These advances have expanded the analytical depth achievable by shotgun proteomics without fundamentally altering its workflow.
Protein Inference and Quantification
Because multiple peptides may originate from the same protein, computational protein inference is required to assemble peptide-level information into protein-level identifications. This step introduces inherent ambiguity, particularly for proteins sharing homologous sequences, and must be handled carefully in shotgun proteomic analysis.
Quantitative shotgun proteomics can be performed using several strategies, including label-free quantification (LFQ), spectral counting, and isobaric labeling approaches such as TMT or iTRAQ. Each quantification approach introduces specific trade-offs between accuracy, missing data, and experimental cost. Consequently, the choice of quantification strategy should align closely with study design and biological questions. (learn more at: Label-based Protein Quantification Technology—iTRAQ, TMT, SILAC)
Biological Interpretation and Pathway Analysis
Once proteins are identified and quantified, shotgun proteomics data are interpreted in a biological context. Differential protein expression analysis, functional annotation, and pathway enrichment are commonly applied to link proteomic changes to cellular processes and phenotypes.
At this stage, integration with complementary omics layers—such as transcriptomics or metabolomics—can reveal regulatory mechanisms that are not apparent from proteomics alone. This integrative perspective has positioned shotgun proteomics as a core component of systems-level biological research.
Advantages and Limitations of Shotgun Proteomics
Shotgun proteomics offers several important advantages that have contributed to its widespread adoption. It enables unbiased, large-scale protein identification across complex samples and is compatible with a wide range of biological systems. The approach is highly scalable and benefits directly from ongoing advances in mass spectrometry instrumentation and bioinformatics.
However, shotgun proteomic analysis also has limitations. Because proteins are inferred from peptides, information about intact proteoforms and combinatorial post-translational modifications may be lost. In addition, data-dependent acquisition can introduce stochastic sampling, leading to missing values across large sample cohorts.
Understanding these strengths and limitations is essential when selecting shotgun proteomics as the appropriate strategy for a given research question.
Applications of Shotgun Proteomics in Biomedical Research
Shotgun proteomics has become a central discovery tool in modern biomedical research due to its ability to provide unbiased, large-scale characterization of complex proteomes. As a core implementation of bottom-up proteomics, shotgun proteomic analysis enables simultaneous identification and relative quantification of thousands of proteins in complex biological samples. This capability has positioned shotgun proteomics as a foundational approach in clinical biomarker discovery, disease mechanism studies, and pharmacological research, where comprehensive proteome coverage is essential for generating mechanistic insight and guiding downstream validation.
CSF Shotgun Proteomics for Neurological Disease Research
One important clinical application of shotgun proteomic analysis is in the study of cerebrospinal fluid (CSF) to identify proteins associated with neurological diseases. CSF is in direct contact with the brain and central nervous system and reflects biochemical changes occurring in those tissues. Untargeted shotgun proteomic analysis allows researchers to profile changes in the CSF proteome, including differential expression of proteins that may serve as biomarkers for disease processes.
In an study on cerebral amyloid angiopathy (CAA), a neurovascular condition related to Alzheimer’s pathology, researchers performed untargeted, data-independent acquisition (DIA) shotgun proteomics on CSF from both an rTg-DI rat model and human participants (sporadic CAA vs controls). The research identified distinct patterns of differentially expressed proteins, including altered levels of proteases (such as cathepsins) and serine protease inhibitors, as well as synaptic proteins that were reduced in patients relative to controls. Importantly, they reported 15 proteins significantly altered in both the rat model and human CAA, including SCG5 and SERPING1, providing a focused starting point for subsequent functional studies and for evaluating the relevance of these proteins in broader neurovascular and neurodegenerative research contexts.
 analysis in rTg-DI models._1767921630_WNo_965d452.webp)
Study design of proteomic cerebrospinal fluid (CSF) analysis in rTg-DI models.
Image reproduced from Vervuurt et al., 2024, Acta Neuropathologica Communications, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
Shotgun Proteomic Analysis in Cancer Biomarker Research
Another major medical application of shotgun proteomics is biomarker discovery in oncology, where researchers use LC-MS/MS to identify proteins that differ between cancerous and non-cancerous tissues or fluids. By profiling these proteomes globally, shotgun proteomic analysis can uncover candidate biomarkers that may serve as early diagnostic indicators or targets for therapeutic intervention.
A recent study used a shotgun proteomic workflow to analyze human pancreatic cancer cell models and patient serum. Through global shotgun proteomic analysis, researchers identified 142 candidate proteins with differential expression in pancreatic cancer relative to normal controls. Subsequent bioinformatic and semi-quantitative analyses related these changes to altered protein networks in cancer cells, and one protein, fibulin-1, emerged as a potential serum biomarker for pancreatic cancer detection due to its significantly lower levels in patient samples. The results provide a basis for subsequent validation studies in independent cohorts and for integration with other molecular and clinical data. Such exploratory analyses contribute to a broader understanding of protein-level changes associated with pancreatic cancer.
Shotgun Proteomics in Pharmacoproteomics and Drug Mechanism Studies
Beyond biomarker discovery, shotgun proteomics is increasingly applied in pharmacoproteomics to investigate how therapeutic compounds modulate global protein expression and cellular pathways. This application is particularly valuable for understanding drug mechanisms of action and adaptive cellular responses.
In a recent study published in Pharmaceutics, researchers treated neuroblastoma cells with two cytoprotective doses of citicoline (0.1 mM and 1 mM) and applied an unbiased shotgun proteomics workflow to quantify global proteome changes over time. The analysis identified and quantified over 4,000 unique proteins per experimental condition (with FDR ≤ 0.01) and found that citicoline induced a pronounced proteome remodeling at 6 hours that diminished by 18 hours, consistent with a transient adaptive response rather than a persistent shift. Pathway clustering of proteins upregulated after treatment highlighted enrichment for mRNA splicing, protein translation, proteostasis regulation via the ubiquitin–proteasome system (UPS), and mitochondrial metabolism following drug exposure. By capturing time-dependent proteome remodeling without prior target selection, this work provides mechanistic insight into drug-induced cellular adaptations relevant to neuroprotective research.
 in SHSY5Y cells identified at 6 h of stimulation with 0.1 mM citicoline or 1 mM citicolines._1767921667_WNo_965d576.webp)
The distribution and overlap of differentially expressed proteins (DEPs) in SHSY5Y cells identified at 6 h of stimulation with 0.1 mM citicoline or 1 mM citicolines.
Image reproduced from Cavaterra et al., 2026, Pharmaceutics, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
Shotgun Proteomics in the Context of Modern Proteomics
As proteomics continues to evolve, shotgun proteomics remains a foundational method that complements newer targeted and top-down approaches. While targeted proteomics excels at precise quantification and top-down proteomics provides proteoform-level resolution, shotgun proteomics offers unmatched breadth and discovery potential. Rather than competing with targeted or top-down proteomics, shotgun proteomics is most often applied in a complementary manner. In many research workflows, global shotgun datasets inform the selection of proteins, peptides, or pathways for subsequent targeted quantification or higher-resolution proteoform analysis. This layered strategy allows researchers to balance discovery, quantitative precision, and molecular detail within a single experimental framework.
Looking ahead, ongoing advances in mass spectrometry instrumentation, chromatographic separation, and computational analysis are expected to further increase the depth, reproducibility, and interpretability of shotgun proteomics data. At the same time, closer integration with other omics modalities and improved experimental standardization will continue to shape how shotgun proteomics contributes to systems biology and translational research. Shotgun proteomics will remain an integral component of proteomic study design, evolving alongside emerging technologies rather than being replaced by them.
Reference
1. Yim YY, Nestler EJ. Cell-Type-Specific Neuroproteomics of Synapses. Biomolecules. 2023 Jun 16;13(6):998. https://doi.org/10.3390/biom13060998
2. Krasny L, Huang PH. Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology. Mol Omics. 2021 Feb 1;17(1):29-42. https://doi.org/10.1039/d0mo00072h
3. Vervuurt M, Schrader JM, de Kort AM, Kersten I, Wessels HJCT, Klijn CJM, Schreuder FHBM, Kuiperij HB, Gloerich J, Van Nostrand WE, Verbeek MM. Cerebrospinal fluid shotgun proteomics identifies distinct proteomic patterns in cerebral amyloid angiopathy rodent models and human patients. Acta Neuropathol Commun. 2024 Jan 8;12(1):6. https://doi.org/10.1186/s40478-023-01698-4
4. Yamamoto T, Boku S, Mitamura K, Taga A. Shotgun Label-Free Proteomic Analysis for Identification of a Potential Diagnostic Biomarker for Pancreatic Cancer. Biomedicines. 2025 Oct 27;13(11):2631. https://doi.org/10.3390/biomedicines13112631
5. Cavaterra D, Giammaria S, Pandino I, Zingale GA, Delli Paoli V, Fiore R, Michelessi M, Roberti G, Carnevale C, Tanga L, et al. Citicoline Triggers Proteome Remodeling and Proteostatic Adaptation: Evidence from Shotgun Proteomics. Pharmaceutics. 2026; 18(1):61. https://doi.org/10.3390/pharmaceutics18010061
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


