An Overview of Single-Cell Spatial Multi-Omics: Advancing Biological Research and Disease Understanding
Single-cell multi-omics is an advanced biotechnology approach that enables the simultaneous analysis of multiple omics layers—such as genomics, transcriptomics, proteomics, and metabolomics—at the single-cell level. This powerful method allows scientists to obtain highly detailed insights into the molecular landscape of cells across different biological states, facilitating a deeper understanding of cellular functions and their roles within complex biological systems. Unlike traditional single-omics analyses, which focus on a single biological layer, multi-omics integrates diverse data types using multimodal techniques, offering a more comprehensive view. This integrated approach unlocks new opportunities for exploring the intricacies of biological systems, revealing their complexity in unprecedented detail.
What is Multi-Omics? Perspectives from the Central Dogma
A fundamental concept in molecular biology is the central dogma, which describes the flow of genetic information: from DNA to RNA, and ultimately from RNA to proteins, which then carry out biological functions. However, this process is not as straightforward as it seems. It is influenced by a variety of intricate regulatory mechanisms, including epigenetic regulation and post-transcriptional modifications, that determine how genetic information is expressed. For example, epigenetic modifications control whether DNA is transcribed into RNA, while post-transcriptional changes influence how RNA is translated into functional proteins. These processes are complex and dynamic, making them ideal for exploration through multi-omics approaches.
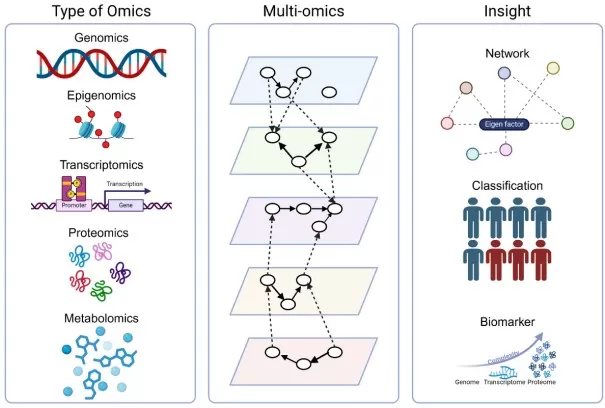
Multi-omics concepts (Hayes et al., 2024)
Multi-omics is an integrated research strategy that allows for a deeper exploration of biological systems by combining data from multiple omics layers—such as genomics, transcriptomics, proteomics, and metabolomics. By analyzing these diverse biological data sets and their interactions, multi-omics provides a more comprehensive and nuanced understanding of cellular processes. The integration of various omics layers enables researchers to synthesize information from multiple angles, uncovering the intricate relationships and regulatory mechanisms that govern biological systems. This approach not only reveals the broader dynamics of biological systems but also provides detailed insights, creating a more holistic and dynamic representation of cellular function.
The Key Domains of Multi-Omics: From Genomics to Metabolomics
Multi-omics integrates key biological fields, including genomics, epigenomics, transcriptomics, proteomics, and metabolomics, with each offering distinct insights into the molecular mechanisms of life. This comprehensive integration provides a holistic understanding of how genes, regulatory processes, proteins, and metabolites interact to drive cellular functions and maintain health.
- Genomics: Genomics studies all genetic information within an organism, focusing on gene structure, function, and interactions.
- Epigenomics: Epigenomics investigates chemical modifications involved in gene expression regulation (e.g., DNA methylation and histone modifications), aiming to uncover cellular epigenetic regulation and explore the mechanisms of life and disease from the source.
- Transcriptomics: Transcriptomics focuses on the complete set of transcripts in a cell or tissue under specific conditions. It studies the process and products of gene transcription, including mRNA and non-coding RNAs. Transcriptomics can reveal which genes are activated or repressed under specific conditions, as well as the expression levels and splicing variations of transcripts, providing insights into gene expression dynamics and regulatory mechanisms.
- Proteomics: Proteomics investigates all proteins expressed in a cell or tissue under specific conditions. Through technologies like mass spectrometry, proteomics identifies proteins, their abundance, and interactions, offering insights into cellular functions.
- Metabolomics: Metabolomics refers to the dynamic collection of endogenous metabolites within an organism. Traditionally, metabolism includes both biosynthesis and biodegradation, and metabolites encompass nucleic acids, proteins, lipids, and other small molecules. Unlike genomics, transcriptomics, and proteomics, metabolomics typically focuses on small molecules with a molecular mass under 1000 Daltons.
Single-Cell Spatial Multi-Omics: Emerging Technologies and Unprecedented Insights
The human body is an incredibly complex system, comprising approximately 37 trillion cells across hundreds of distinct cell types. Despite originating from a single fertilized egg, these cells accumulate genetic and epigenetic variations during processes like regeneration and differentiation, leading to significant cellular heterogeneity within the same tissue, organ, or cell type. This heterogeneity means that different cells can have varying roles in physiological and pathological processes. Understanding this variation is critical for advancing both basic and clinical research.
Traditional bulk omics analysis assesses the overall status of a sample—such as a tissue block or cell population—but it cannot distinguish the differences between individual cells. In contrast, single-cell data offers insights into cellular heterogeneity, while spatial omics enhances this by providing information on the spatial organization and proximity of cells within tissues. This combination of technologies enables more precise and detailed analysis of biological systems, allowing researchers to construct high-resolution multi-omics cell atlases and create digital models of life itself.
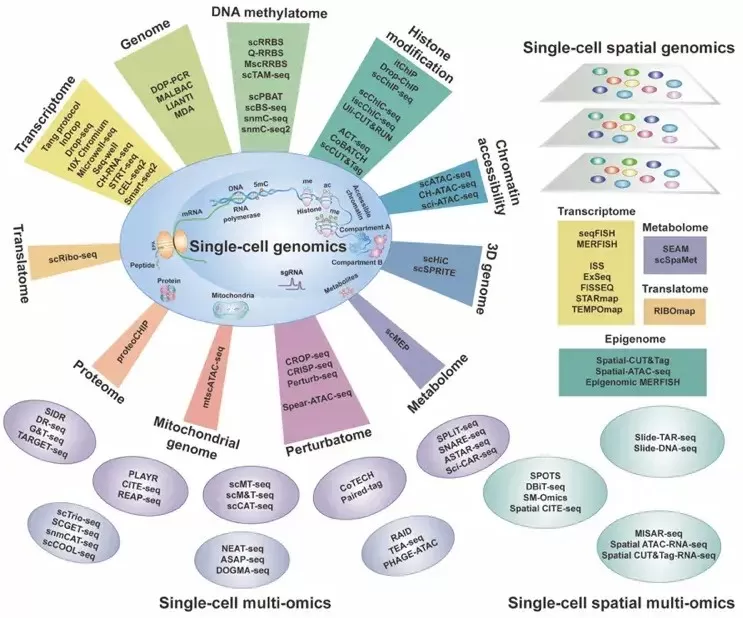
Single-cell Multi-omics Technologies (Wang et al., 2025)
Single-Cell Sequencing Technologies
Single-cell sequencing has evolved significantly, expanding from early transcriptome sequencing to include whole-genome, epigenome, and proteome sequencing. What began as single-omics has now advanced to multi-omics approaches, from single-cell sequencing to subcellular sequencing, and from analyzing dozens of cells to studying millions. This technology uncovers the inherent heterogeneity between individual cells, providing a more accurate and comprehensive understanding of biological processes. For instance, single-cell RNA sequencing (RNA-seq) allows for detailed analysis of gene expression in each individual cell, offering valuable insights into cellular diversity, cell fate determination, and cellular behavior in specific pathological conditions.
Spatial Transcriptomics
Spatial omics technologies add another layer of depth by providing spatial coordinates at the single-cell level. This enables the mapping of cells within their tissue context, revealing their location and the interactions between cells. Techniques such as spatial in situ hybridization (SISH), spatial in situ sequencing (SISS), spatial in situ microdissection (SISM), and spatial in situ labeling (SISB) are key to achieving this. The integration of spatial data with multi-omics information offers complementary insights, providing a more complete picture of cellular functions and their role in tissue architecture.
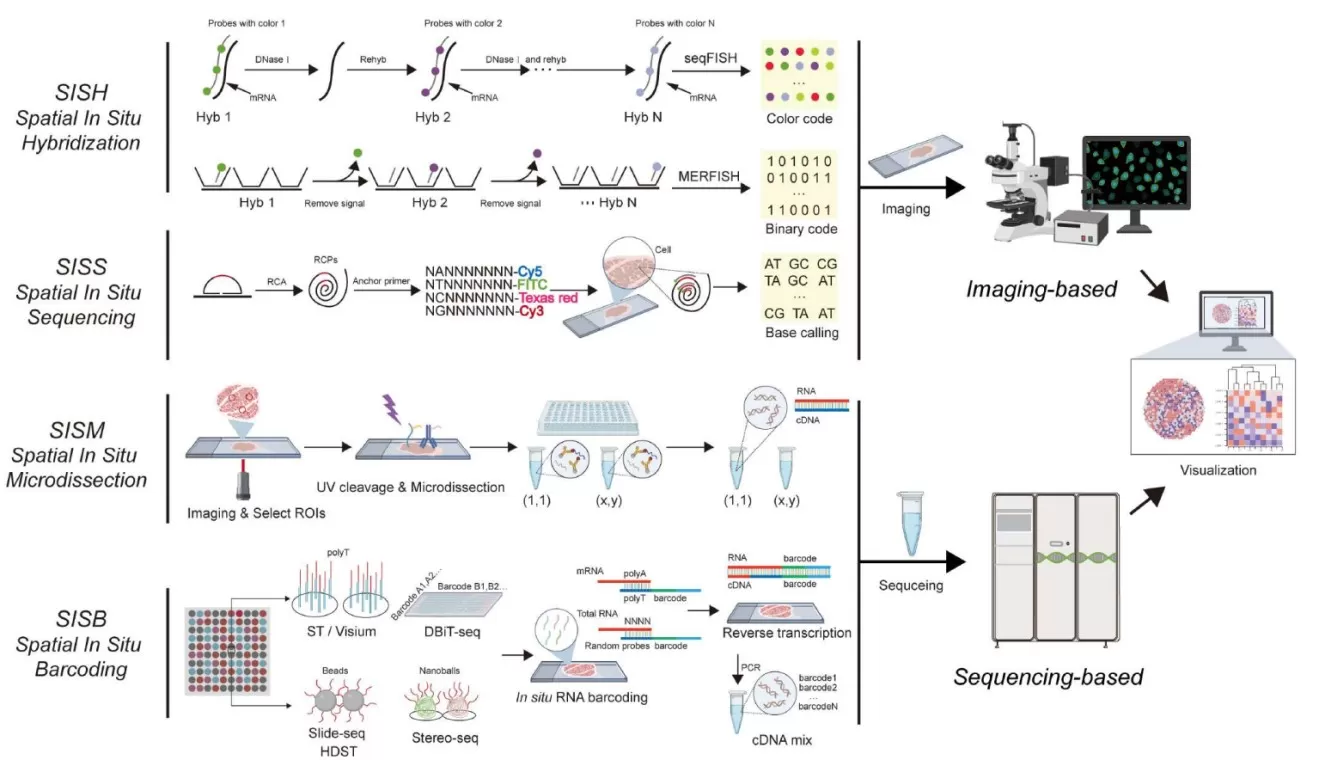
Overview of the four main spatial transcriptomics methods (Wang et al., 2025)
Single-Cell and Spatial Mass Spectrometry Technologies
Mass spectrometry applied to single-cell and spatial analysis allows for the high-precision, high-throughput investigation of biomolecules like proteins and metabolites. This method reveals the biochemical properties and functional states of individual cells, providing direct visualization of molecular diversity within cell populations. By offering insights into the molecular landscape of cells, this technique is essential for understanding cellular complexity, elucidating cellular functions, and investigating disease mechanisms at the single-cell level.
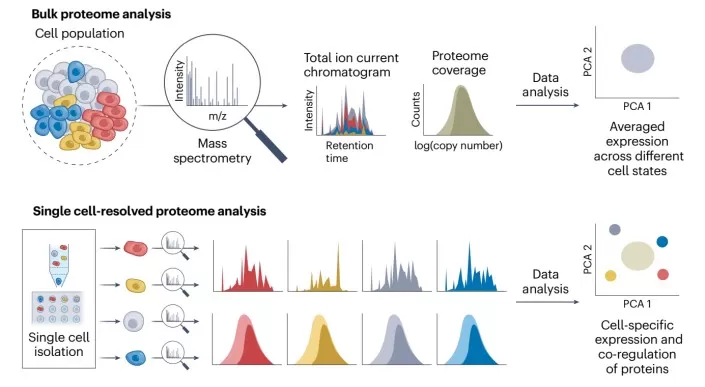
Deep single-cell proteomics to gain biological insights (Sinn and Demichev, 2025)
The Future of Single-Cell Multi-Omics: Transforming Research and Healthcare
Recent advancements in multi-omics technologies, which integrate data from genomics, transcriptomics, proteomics, and metabolomics, are transforming biological research by providing a more comprehensive view of cellular processes. Single-cell spatial multi-omics takes this a step further by enabling the study of individual cells in their native tissue environments, uncovering cellular diversity and interactions that were previously hidden. This approach allows researchers to map the spatial organization of cells, offering critical insights into how diseases like cancer progress, metastasize, and develop resistance to treatment. By examining how cells interact within their tissue context, single-cell spatial omics opens new pathways for understanding disease mechanisms and identifying potential therapeutic targets.
Looking ahead, the applications of single-cell spatial multi-omics in clinical settings are poised to be transformative. In diagnostics, these technologies will enable the creation of detailed molecular profiles of patient tissues, leading to earlier, more accurate diagnoses and better disease stratification. By understanding how different cell types within a tissue respond to disease or treatment, clinicians can make more informed decisions about personalized treatment plans. In drug development, spatial multi-omics will facilitate the identification of biomarkers and therapeutic targets, allowing for more precise testing of drugs and accelerating the development of personalized therapies. As these technologies continue to evolve, they promise to revolutionize the way we approach disease diagnosis, treatment, and prevention, ultimately paving the way for more targeted, effective healthcare solutions.
Reference
1. Hayes CN, Nakahara H, Ono A, Tsuge M, Oka S. From Omics to Multi-Omics: A Review of Advantages and Tradeoffs. Genes (Basel). 2024;15(12):1551. Published 2024 Nov 29. doi:10.3390/genes15121551
2. Wang J, Ye F, Chai H, et al. Advances and applications in single-cell and spatial genomics. Sci China Life Sci. 2025;68(5):1226-1282. doi:10.1007/s11427-024-2770-x
3. Sinn LR, Demichev V. Entering the era of deep single-cell proteomics. Nat Methods. 2025;22(3):459-460. doi:10.1038/s41592-025-02620-7
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


