Spatial Metabolomics with MALDI-MSI: Workflow & Applications
Spatial metabolomics is an emerging analytical approach that visualizes the distribution of metabolites within tissues or cells, allowing researchers to understand how metabolic activity varies across different microenvironments. Among the most impactful technologies enabling this field is Matrix‑Assisted Laser Desorption/Ionization Mass Spectrometry Imaging (MALDI‑MSI). Unlike conventional metabolomics, which provides averaged metabolite profiles, MALDI‑MSI preserves spatial information, making it possible to determine where metabolites accumulate rather than only how much is present. This expanded overview introduces the MALDI‑MSI workflow and highlights its growing applications across biology and medicine.
Workflow of MALDI-MSI Spatial Metabolomics
The MALDI‑MSI workflow typically consists of four essential steps. Each stage plays a critical role in ensuring accurate spatial resolution and reliable metabolite detection.
1. Sample Preparation
Tissue Sectioning: Fresh or frozen tissues are sliced into thin sections, usually 10–15 µm thick, using a cryostat for frozen samples or a microtome for fixed samples. These thin sections preserve the tissue’s structural integrity and help maintain native metabolite distribution.
Matrix Application: A suitable chemical matrix—such as DHB for lipids or CHCA for peptides—is uniformly sprayed or sublimated onto the tissue surface. The matrix enables efficient desorption and ionization of metabolites when exposed to the laser beam.
Quality Control: Staining methods like hematoxylin and eosin (H&E) are often used to evaluate tissue morphology. These reference images assist with aligning histological features to the later MSI-derived metabolic maps.
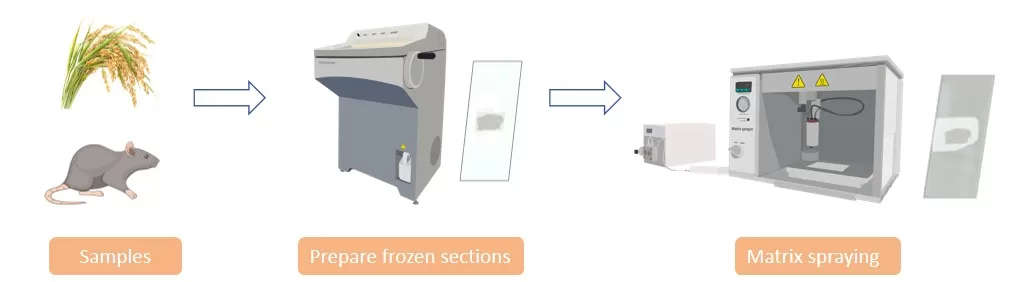
Sample Preparation for Spatial Omics
2. Mass Spectrometry Imaging
In this step, the prepared tissue section is loaded into the MALDI-MS instrument. A precise laser beam scans across the sample in a raster pattern, ionizing molecules at each predefined pixel. The mass spectrometer then records the mass-to-charge ratio (m/z) of ions produced at every pixel position. Modern instruments achieve resolutions of 10–50 µm, enabling visualization of fine structural details.

MALDI-Mass Spectrometry Imaging
3. Data Acquisition
High-resolution mass spectra are collected throughout the tissue section. Many systems incorporate tandem mass spectrometry (MS/MS), which fragments selected ions to confirm their chemical identity. These fragmentation patterns are matched to public databases or in-house database, enhancing the reliability of metabolite identification.
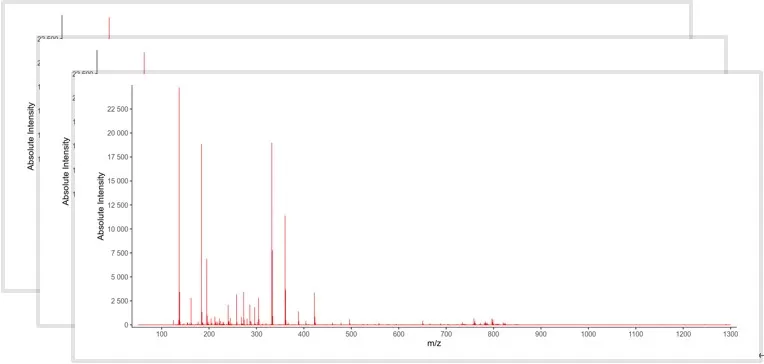
Data collection for MSI
4. Data Analysis
Preprocessing: Raw spectra are normalized to correct for varying ionization efficiency and aligned to account for any spatial shifts.
Metabolite Identification: Detected ions are compared with reference databases using both accurate mass and MS/MS fragmentation patterns.
Spatial Mapping: Analysis software—such as SCiLS Lab or ImageJ—generates heatmaps indicating spatial variation in metabolite levels. Advanced statistical tools including PCA and t‑SNE further assist in revealing metabolic gradients and identifying correlations with specific tissue structures.
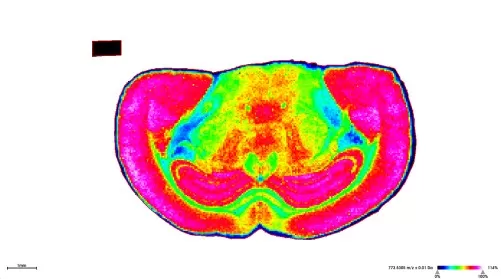
Heatmaps of spatial variation in metabolite levels
Applications of MALDI-MSI Spatial Metabolomics
The ability to associate metabolite localization with biological function has enabled MALDI‑MSI to contribute significantly to several research areas.
1. Cancer Research
Metabolic Heterogeneity: MALDI‑MSI reveals variations in metabolic pathways across tumor regions. For example, elevated lactate levels in hypoxic tumor cores reflect intensified glycolysis, which often contributes to tumor progression and treatment resistance.
Biomarker Discovery: Spatially resolved metabolite profiles—such as distinctive phospholipid signatures—can support tumor classification and help predict patient prognosis.
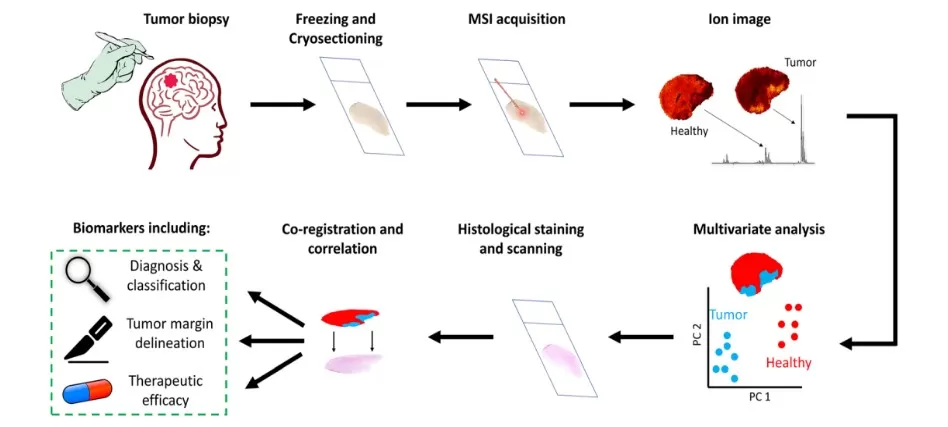
MSI spatial metabolomics for cancer research
2. Neuroscience
Brain Metabolism: MALDI‑MSI allows visualization of neurotransmitters and lipid species across brain regions, providing insights into how metabolic networks relate to neural function. For instance, mapping acetylcholine distribution in the hippocampus yields valuable clues about learning and memory.
Neurodegenerative Disease Research: This technique detects spatial changes in key molecules associated with diseases like Alzheimer’s and Parkinson’s, offering a clearer understanding of disease progression and affected regions.
3. Drug Development
Pharmacokinetics: MALDI‑MSI provides direct visualization of drug molecules and their metabolites in tissues, improving understanding of drug absorption, distribution, and clearance.
Target Engagement: By mapping drug-related ions to specific tissue compartments, researchers can confirm whether therapeutic compounds effectively reach their intended cellular or subcellular targets.
Conclusion: Future Directions for MALDI-MSI Spatial Metabolomics
MALDI‑MSI has revolutionized spatial metabolomics by providing an unprecedented view of metabolite localization within biological tissues. Its ability to bridge chemical identity with spatial context has advanced fields ranging from oncology to neuroscience and pharmacology. As instrumentation and data analysis tools continue to improve, MALDI‑MSI will play an increasingly vital role in uncovering complex biological mechanisms and guiding the development of precision medicine.
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


