Spatial Multi-Omics Integration: Technologies, Data Fusion and Biological Applications
Why “Spatial Multi-Omics” Has Become a Leading Keyword in Top Journals
Spatial multi-omics refers to a suite of technologies that capture multimodal information from the transcriptome, proteome, and metabolome while preserving tissue architecture and cellular niche–level resolution. These approaches integrate multi-omics signals through computational algorithms, enabling a clear understanding of ‘which cells reside where’ and ‘how they interact’—a major limitation of traditional ‘grind-and-extract’ bulk omics workflows.
As one of the most transformative directions in contemporary life sciences, spatial multi-omics combines spatial transcriptomics, spatial proteomics, and spatial metabolomics to decode tissue organization, cellular functional states, and interaction mechanisms without compromising structural context. Owing to its exceptional resolution of cellular microenvironments, spatial multi-omics is reshaping the way we understand complex biological systems, while integrated spatial omics pipelines are rapidly accelerating progress in precision medicine.
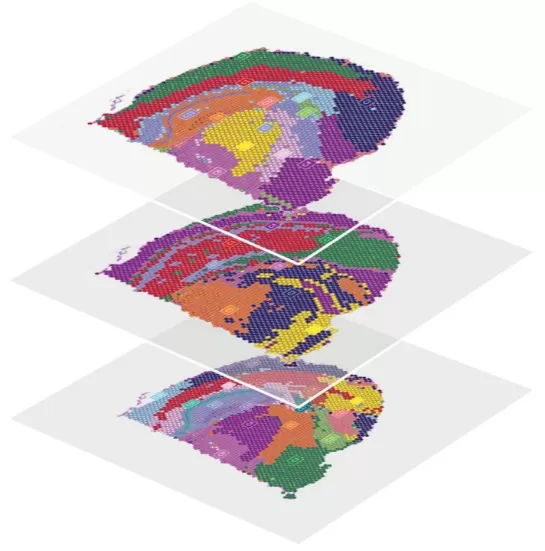
Spatial Multi-Omics Integration
Major Modalities and Leading Platforms in Spatial Multi-Omics
Modern spatial multi-omics research is supported by three core technological pillars: spatial transcriptomics, spatial proteomics, and spatial metabolomics. Using multimodal imaging strategies, these methods provide spatially resolved information at the RNA, protein, and metabolite levels, collectively generating a comprehensive molecular atlas.
Comparison of the most widely used platforms across these modalities
|
Omics Type |
Technical Principle |
Representative Platform |
Resolution |
Key Advantages |
|
Spatial Transcriptomics |
Sequencing-based |
10x Visium |
55 μm |
Commercially mature with stable data quality |
|
Visium CytAssist |
55 μm |
Compatible with standard microscope slides, high flexibility |
||
|
10x Visium HD |
2 μm |
Higher resolution with cell-level localization |
||
|
Stereo-seq |
0.5 μm |
Currently the highest resolution, nanoscale precision |
||
|
Xenium |
Single-cell |
Single-cell resolution with a fully commercialized solution |
||
|
Spatial Proteomics |
Mass spectrometry imaging |
Imaging Mass Cytometry |
Single-cell/subcellular |
High marker count with accurate quantification |
|
MIBI-TOF |
1 μm |
High multiplexing capability for deep phenotyping |
||
|
Fluorescence imaging |
CODEX/PhenoCycler |
Single-cell |
High-dimensional phenotyping with preserved tissue structure |
|
|
Multiplexed IF (mIF) |
Single-cell |
Widely available and technically mature |
||
|
Cell DIVE |
Single-cell |
Ultra-high-dimensional imaging with flexible marker combinations |
||
|
Mass spectrometry imaging |
MALDI-MSI |
5–100 μm |
Label-free detection with broad metabolite coverage |
|
|
DESI-MSI |
50–200 μm |
Ambient detection with minimal sample damage |
||
|
AFADESI-MSI |
20–100 μm |
High sensitivity in atmospheric conditions |
||
|
3D DESI |
50–200 μm |
Capability for three-dimensional spatial reconstruction |
Choosing the appropriate platform is a crucial first step in designing a spatial omics workflow. This choice requires careful balancing of omics coverage, spatial resolution, sensitivity, and compatibility with downstream analysis.
Challenges in Integrating Spatial Multi-Omics Data
Integrating spatial multi-omics data presents several technical challenges. Different omics technologies produce datasets with distinct resolutions, sensitivity ranges, and noise characteristics. For instance, spatial metabolomics typically offers lower spatial resolution; spatial proteomic imaging achieves subcellular resolution but detects a relatively small number of proteins; and microdissection-based methods may lack precise spatial coordinates. These inconsistencies make spatial data fusion difficult—particularly for coordinate co-localization and cross-omics harmonization.
Key challenges include:
(1) mismatched spatial resolution across platforms;
(2) variations in technical noise;
(3) batch-effect correction across experiments;
(4) the computational burden of large-scale data processing.
To address these issues, current approaches employ deep learning–based cross-modal alignment, probabilistic graphical models, and advanced statistical frameworks that enable robust integration across multiple modalities with heterogeneous data quality.
Advanced Technologies for Spatial Registration and Data Fusion
Spatial registration is a central step in achieving accurate integration of spatial multi-omics datasets. Using advanced computer vision techniques and deep learning algorithms, researchers can precisely align tissue sections generated from different platforms, ensuring spatial correspondence across modalities.
Spatial Alignment of Multimodal Data
|
Method / Tool |
Core Strategy |
Supported Modalities |
Typical Scenario |
|
PASTE-C² |
Probabilistic coupling + elastic deformation |
Transcriptome + metabolome + proteome |
Consecutive sections |
|
MAGPIE |
Proxy-slice indirect registration |
Transcriptome + metabolome + proteome |
Consecutive sections |
|
STaCker |
Deep-learning U-Net deformation |
Transcriptome + proteome |
Same section |
|
SpaMosaic |
Graph neural network with contrastive learning |
Any three modalities |
Mosaic integration |
|
VoltRon |
Image features (SIFT/ORB) |
Transcriptome + proteome |
Imaging–sequencing alignment |
Breakthrough Biological Applications and Case Studies
Spatial multi-omics has driven major breakthroughs across diverse biomedical disciplines. Representative studies include:
1. Tumor Immune Microenvironment
Ravi et al. (2022, Cancer Cell, DOI:10.1016/j.ccell.2022.05.009) applied spatial transcriptomics, metabolomics, and proteomics to characterize glioblastoma. By identifying transcriptional programs shared across patients, they revealed that glioblastoma exhibits spatial segregation based on lineage state and adapts to inflammatory or metabolic cues. Integrated spatial proteomic and metabolomic analyses further uncovered locally dependent tumor–host interactions, giving rise to spatially restricted adaptive transcriptional programs.
2. Neurodegenerative Disease Mechanisms
Lee et al. (2023, Cell Reports, DOI: 10.1016/j.celrep.2023.112196) used spatial multi-omics to highlight the central regulatory role of APOE in microglial immunometabolism and provided an interactive resource to facilitate discovery and validation in neurodegeneration research.
3. Developmental Biology
The Lengyel group at the University of Chicago (2025, Cancer Cell, DOI:10.1016/j.ccell.2025.06.004) combined spatial proteomics and transcriptomics to delineate the evolutionary progression from SBT to LGSC and to map the metastatic spread across stromal and tumor compartments.
Leading Analytical Tools and Computational Methods
A range of specialized analytical tools has been developed to process spatial multi-omics datasets. Among the top-performing platforms are:
- Seurat v5 — Supports multimodal spatial integration; one of the most widely used frameworks with over 100,000 monthly downloads.
- Squidpy — Focuses on spatial interaction network analysis and excels at pathway co-localization studies.
- Giotto Suite — Provides an end-to-end analysis pipeline supporting 50+ spatial omics modalities.
- MISTY — Designed for multiscale feature integration and spatial data fusion, widely applied in tumor microenvironment research.
These tools leverage advanced machine learning and statistical modeling, greatly enhancing the ability to extract biological insights from complex spatial datasets. According to a 2023 ‘Nature Biotechnology’ benchmark study, Seurat and Squidpy ranked highest in overall analytical performance. In practice, the most powerful strategies often integrate multiple tools to progress from raw image data to biological discovery.
Read more
- Spatial Metabolomics: MALDI-MSI vs. AFADESI-MSI, Key Technologies, and Applications
- MALDI, DESI, or SIMS? How to Choose the Best MSI Techniques for Spatial Metabolomics
- Is Your Spatial Metabolomics Sample Ready?
- How to Prepare Samples for Spatial Metabolomics: The Essential Guide You Need
- Top FAQs on Spatial Metabolomics Sample Preparation
- Unlocking the Key Parameters of Spatial Metabolomics: Decoding Precision Metabolic Mapping
- Spatial Metabolomics Resolution Guide: From 100μm to 5μm in Plant Tissue Imaging
- Advanced Data Analysis in Spatial Metabolomics
- Spatial Metabolomics in Cancer Research: Unlocking the Metabolic Code for Precision Oncology
- Tracking Drugs in 4D: The Future of Pharmacokinetics with Spatial Metabolomics
- Spatial Metabolomics: Transforming Biomedical and Agricultural Research
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


