Decoding Fatty Liver Disease: A Spatial Multi-Omics Breakthrough
We are excited to feature a groundbreaking study recently published in ‘Nature Genetics’, "Spatially resolved multi-omics of human metabolic dysfunction-associated steatotic liver disease." This research leverages an advanced multi-omics approach, including single-cell RNA sequencing, spatial transcriptomics, and spatial metabolomics using Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging (MALDI-MSI), a core service at MetwareBio. Analyzing 61 human livers across disease stages, the study aimed to construct a high-resolution spatial atlas of MASLD, uncovering novel cellular regulators and metabolic dysfunctions driving disease progression.
Metabolic dysfunction-associated steatotic liver disease (MASLD) has become a leading cause of chronic liver disease globally, progressing from simple steatosis (MASL) to steatohepatitis (MASH), which can lead to cirrhosis and liver cancer. Understanding the complex cellular and molecular interplay within the liver's structured tissue environment is crucial for developing effective treatments. Traditional single-cell analyses, while powerful, lack spatial context. This study underscores the necessity of spatially resolved technologies—like spatial transcriptomics and metabolomics—to capture the region-specific gene expression and metabolic alterations that define MASLD pathogenesis.
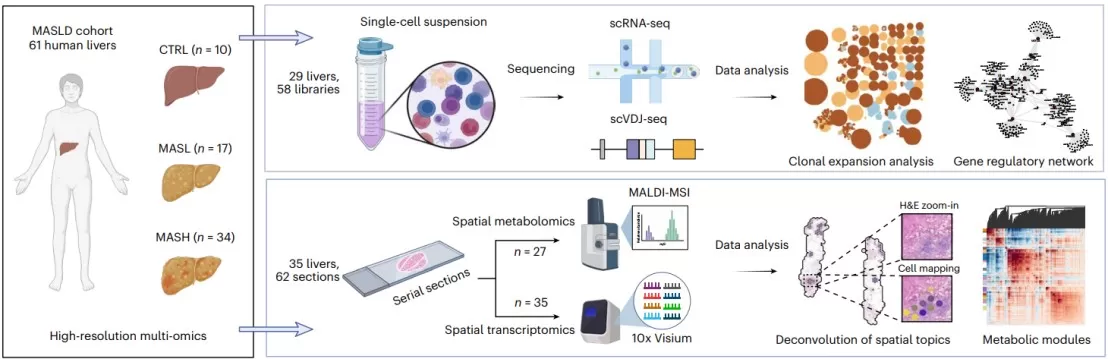
Spatial multi-omics for human MASLD atlas
MITF Emerges as a Key Regulator in Lipid-Handling Macrophages
Using single-cell RNA sequencing and gene regulatory network analysis (pySCENIC), the researchers identified the microphthalmia-associated transcription factor (MITF) as a master regulator specific to lipid-associated macrophages (LAMs). MITF activity was elevated in MASH and correlated with a lipid-handling phenotype. Functional experiments in THP-1 cells confirmed that MITF overexpression upregulates key LAM markers and genes involved in fatty acid oxidation, mediated through the PGC1α-PPARγ signaling axis. This reveals a previously unknown mechanism where MITF drives metabolic reprogramming in LAMs, enhancing their lipid-processing capacity.
 plot of cells_1764039551_WNo_965d532.webp)
Uniform manifold approximation and projection (UMAP) plot of cells
LAMs Exhibit a Hepatoprotective Role via HGF Secretion
Intercellular communication analysis (FlowSig) identified a significant increase in hepatocyte growth factor (HGF) outflow from LAMs in advanced MASH. RNAscope and immunofluorescence validated HGF expression in TREM2+ LAMs. Conditioned medium from MITF-overexpressing THP-1 cells increased HGF production, promoting hepatocyte proliferation and reducing apoptosis in vitro. This suggests LAMs play a protective role in MASLD by secreting HGF, potentially mitigating liver injury through the HGF-MET axis—a finding that shifts the perception of LAMs from solely pathogenic to also reparative.
Spatial Topic Modeling Uncovers a Fibrosis-Associated Gene Program
The team applied STAMP (Spatial Transcriptomics Analysis with Topic Modeling) to deconvolute spatial transcriptomics data, identifying a fibrosis-associated "Topic5" enriched in advanced MASH. This topic included collagen and fibrosis-related genes (e.g., COL1A1, THY1) and spatially aligned with fibrotic regions, hepatic stellate cells (HSCs), and central vein endothelial cells. Cell-cell interaction analysis (CellPhoneDB) predicted a pro-fibrotic RSPO3–LGR6 ligand-receptor pair between these cell types, suggesting a new signaling axis driving liver fibrogenesis.
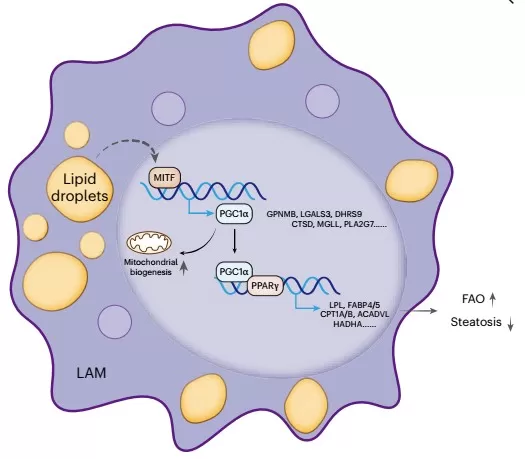
LAMs exert hepatoprotective function via the HGF–MET axis
Spatial Metabolomics Reveals MASLD-Specific Phospholipid Accumulation
Through MALDI-MSI-based spatial metabolomics and Hotspot analysis, the study identified disease-specific metabolic modules. Module 5, strongly correlated with MASLD severity, was enriched in phospholipids (e.g., phosphatidic acid, phosphatidylethanolamine) containing very-long-chain fatty acids. Integration with spatial transcriptomics showed co-localization with LAM-rich and fibrotic regions. Notably, the LAM-specific gene PLA2G7 (encoding lipoprotein-associated phospholipase A2) was linked to this phospholipid metabolism, suggesting a role in modulating ferroptosis resistance and sustaining LAM function in fatty livers.
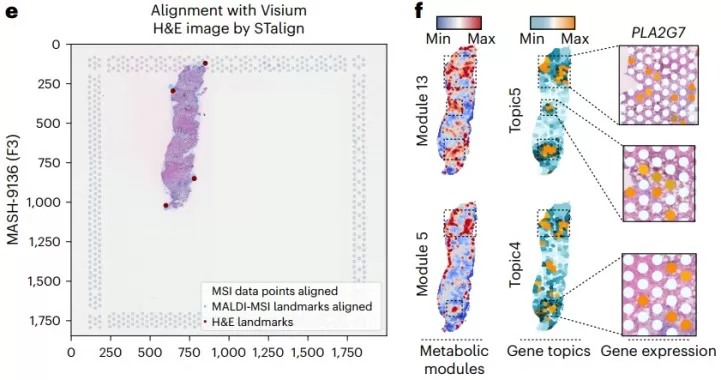
MSI data points and H&E image alignment and Spatial distribution of Metabolic modules
This study provides an unprecedented spatially resolved multi-omics atlas of human MASLD, highlighting MITF as a novel regulator of LAM lipid metabolism and revealing their dual role in disease progression and protection. The integration of spatial transcriptomics and metabolomics was pivotal in linking region-specific gene programs with metabolic alterations, offering a holistic view of MASLD pathology. Technologies like MALDI-MSI spatial metabolomics are indispensable for elucidating the complex metabolic dysfunctions in chronic diseases, presenting new avenues for biomarker discovery and targeted therapies. The insights gained pave the way for similar multi-omics applications in other complex tissues and diseases, promising to accelerate the translation of spatial biology into clinical practice.
At MetWare Bio, we specialize in providing cutting-edge spatial metabolomics services, including MALDI-MSI, to empower groundbreaking research like this. Our platform offers high-sensitivity, spatially resolved metabolite profiling directly from tissue sections, enabling you to uncover critical metabolic insights in their native tissue context. With a team of expert scientists, state-of-the-art mass spectrometry technology, and a commitment to delivering end-to-end support—from experimental design to data analysis—we ensure robust and reproducible results. Contact us now to explore how our spatial multi-omics solutions can advance your research.
Reference
Li, Z., Luo, G., Gan, C. et al. Spatially resolved multi-omics of human metabolic dysfunction-associated steatotic liver disease. Nat Genet (2025). https://doi.org/10.1038/s41588-025-02407-8
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


