Spatial Omics Technologies: Classification of Transcriptomics, Proteomics and Metabolomics
What Is Spatial Omics?
Spatial omics refers to a suite of emerging technologies aimed at performing high-throughput, high-resolution ‘in situ’ detection and analysis of biomolecules—such as RNA, DNA, proteins, and metabolites—while preserving the original spatial location of tissues or cells. In essence, it addresses key questions that traditional omics cannot: “Where are these molecules or cells located?” and “How do they interact with neighboring cells or structures?” Its major features include:
In Situ Analysis: Analyses are performed directly on tissue sections without disrupting tissue architecture, enabling the acquisition of “GPS-like coordinates” of molecular distribution and preserving the sample’s native spatial relationships and microenvironment.
High Spatial Resolution for Creating “Molecular Maps”: These technologies can generate detailed distribution maps at subcellular, single-cell, or near–single-cell resolution, revealing molecular spatial heterogeneity and elucidating the spatial dynamics of gene, protein, and metabolite expression in tumor microenvironments, developmental systems, or structurally diverse plant tissues.
High Throughput: A single experiment can simultaneously detect thousands of genes, proteins, or metabolites—producing millions of data points from one experiment and one tissue section—and effectively moves beyond the era of “one marker per gel.”
Multi-Omics Coverage: In addition to gene expression (spatial transcriptomics), spatial omics encompasses protein accumulation (spatial proteomics) and metabolite distribution (spatial metabolomics), forming a multidimensional, integrated multi-omics perspective.
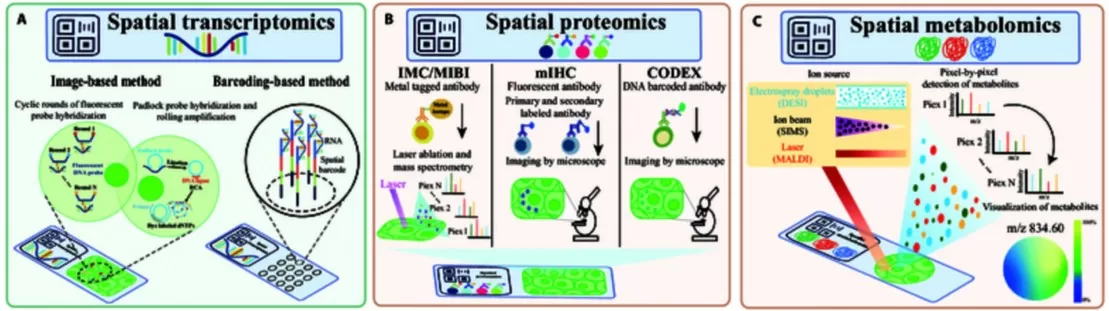
Spatial Omics Technologies
Spatial Omics vs. Traditional Omics
Traditional biological studies often rely on homogenizing cells or tissues into uniform samples for molecular analysis. Although such methods yield abundant molecular information, they cannot pinpoint the precise spatial distribution of these molecules within an organism. As a result, molecular data alone are insufficient for fully understanding tissue development or complex diseases.
Spatial omics addresses this limitation by linking molecular functions to spatial context, thereby overcoming the constraints of traditional omics with respect to resolution, dynamic observation, and tissue structural interpretation. It holds significant promise, especially in oncology, neuroscience, and developmental biology.
Spatial Omics Technologies
The emergence of spatial omics signifies a new era of “spatial decoding” in systems biology and biomedical research. This advancement relies heavily on high-throughput sequencing, high-resolution imaging, and AI-based three-dimensional spatial reconstruction. These innovations not only resolve the long-standing issue of “seeing the forest but not the trees” but also enable multidimensional, high-precision mapping from molecular expression to cell–cell interactions.
Each technological advancement reinforces the paradigm of “technology-driven discovery.” In this evolving field, choosing the optimal technology for specific biological challenges—and understanding each method’s advantages and limitations—is essential.
Spatial Transcriptomics
Spatial transcriptomics methods fall into four major categories:
1. Spatial Isolation or Microdissection (SISM)
Representative technologies: Tomo-seq, DSP, GEO-seq
Specific tissue regions are physically isolated or labeled for subsequent DNA or RNA extraction and analysis.
2. Spatial In Situ Hybridization (SISH)
Representative technologies: seqFISH, MERFISH, STARmap
Labeled probes are applied directly to tissue sections to capture the spatial positions of specific RNA molecules along with sequence information.
3. Spatial In Situ Sequencing (SISS)
Representative technologies: FISSEQ, 10x Xenium
Sequencing is performed directly on fixed tissue sections.
4. Spatial Barcoding (SISB)
Representative technologies: Slide-seq, DBiT-seq, 10x Visium, Stereo-seq, Seq-Scope
mRNA from defined spatial regions is linked to specific DNA barcodes, enabling reconstruction of spatial expression patterns after bulk sequencing.
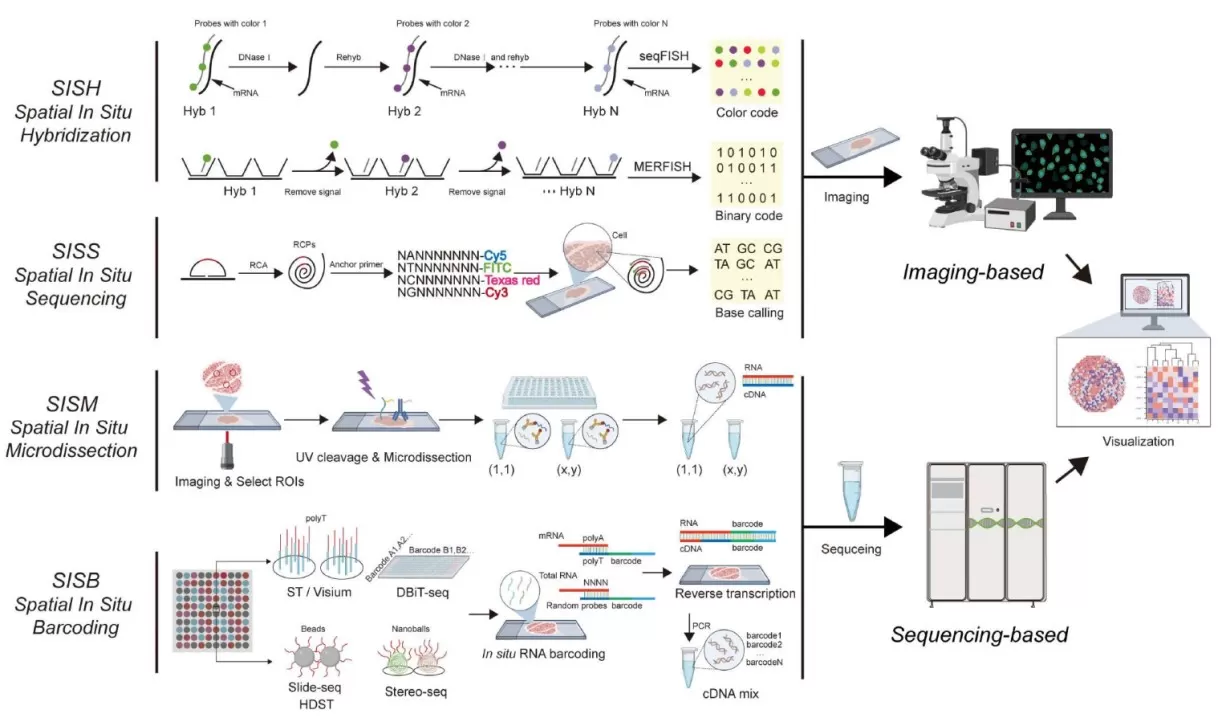
Types of Spatial Transcriptomics Technologies
Currently, spatial barcoding platforms such as 10x Visium and BGI Stereo-seq are widely adopted. Both utilize arrays of DNA oligonucleotide capture probes with poly(T) sequences to bind mRNA. Captured transcripts receive spatial barcodes and UMIs for subsequent localization and quantification. This strategy reduces dependence on complex imaging systems and supports rapid, high-throughput profiling of spatial gene expression.
_1763694057_WNo_800d499.webp)
Spatial Barcode Method (Bressan et al. Science. 2023)
Spatial Proteomics
Subcellular protein localization is tightly regulated and strongly tied to protein function. Therefore, mapping the spatial proteome—protein distribution and dynamics at subcellular resolution—is crucial for understanding cell biology. Spatial proteomics integrates precise cell identification, accurate microdissection, high-recovery sample transfer, and highly sensitive sample preparation. Advanced tools such as multiplex in situ imaging and the SISPROT ultrasensitive processing platform support systematic localization, qualitative assessment, and quantitative analysis of specific microenvironments—such as single-cell spatial positions and tissue structures—within pathological sections.
Spatially resolved proteomics provides functional insights distinct from spatial transcriptomics. Current in situ proteomic strategies fall into two main categories:
1. Antibody-Based Multiplex Imaging
Evolving from traditional IHC/IF, these methods enable high-plex imaging via cyclic staining or multiplexed labeling. Representative platforms include cycIF, CODEX, IBEX, MIBI-TOF and IMC. They allow simultaneous localization of dozens of proteins within a single tissue section at subcellular resolution, with IMC and CODEX capable of detecting over 50 proteins at single-cell resolution. However, these approaches face challenges such as spectral overlap, tissue autofluorescence, and lengthy multi-cycle imaging. DNA-barcoded and metal-tagged antibodies have been introduced to enhance sensitivity and throughput. Technologies such as Spatial CITE-seq and metal isotope–based MIBI/IMC further expand multiplexing capacity.
2. Mass Spectrometry Imaging (MSI) and Microdissection
Antibody-free approaches directly detect protein distributions using mass spectrometry. Representative MSI techniques include: MALDI-MSI and SIMS. These methods allow direct detection of peptides and higher-molecular-weight proteins, including histone modifications. Laser microdissection (LMD) coupled with highly sensitive LC–MS enables deep proteomic profiling of specific tissue regions or even single cells. Recent technological improvements now allow identification of 4,000–6,000 proteins from approximately 50 cells. In the near future, spatial proteomics capable of profiling tens of thousands of proteins in small tissue domains is anticipated.
A notable example is Deep Visual Proteomics (DVP), which integrates high-resolution microscopy, AI image analysis, laser dissection, and deep MS to quantify thousands of proteins and localize them to cellular or subcellular structures within a single FFPE section. Spatial proteomics was recognized as Nature Methods’ 2024 Method of the Year, signifying its rapid maturation and growing scientific impact.
_1763694128_WNo_743d547.webp)
Spatially Resolved Tissue Proteomics Techniques (Liotta et al. Expert Rev Proteomics. 2021)
Spatial Metabolomics
Metabolites define organismal phenotype and offer direct insights into biological processes and mechanisms. Although chromatography–mass spectrometry is powerful for metabolite analysis, the homogenization step eliminates spatial information. Spatial metabolomics primarily employs mass spectrometry imaging (MSI), which integrates mass spectrometry with two-dimensional spatial mapping to achieve both qualitative and quantitative metabolite detection. MSI is broadly applied to characterize the levels and spatial distribution of endogenous metabolites and exogenous drugs in plant and animal tissues.
Compared with fluorescence or radiolabel imaging, MSI requires no chemical or radioactive labeling, minimal sample preparation, and provides high sensitivity, rapid analysis, and retention of spatial information.

Principles of Mass Spectrometry Imaging Technology
MSI is generally classified into four types based on ionization source:
A matrix is applied to the sample surface, and laser irradiation causes matrix–analyte co-crystals to ionize and enter the mass spectrometer. It is suitable for large molecules (proteins) and small molecules, with high sensitivity enabling detection of low-abundance metabolites.
2. DESI-MSI
An ionized solvent spray desorbs and ionizes analytes directly from the sample surface. It requires minimal sample preparation, no matrix, is non-destructive, and is compatible with fresh tissues. It is suitable for lipids, metabolites, drug molecules, and clinical samples.
3. LAESI-MSI
A mid-infrared laser excites water molecules within the sample, causing evaporation and partial ablation. The ablated material is captured by electrospray and ionized for MS analysis. It offers simple sample preparation and high sensitivity without requiring a matrix, though spatial resolution is lower. Samples must contain water and be sufficiently stable.
4. SIMS-MSI
A high-energy primary ion beam bombards the sample surface, sputtering secondary ions for MS detection. It provides extremely high spatial resolution and is suitable for elements, isotopes, small metabolites, fragmented organic molecules, and surface contaminants.
Learn more at: MALDI, DESI, or SIMS? How to Choose the Best MSI Techniques for Spatial Metabolomics
Conclusion: Future Directions for Spatial Omics Technologies
Spatial omics represents the forefront of life sciences. By integrating genomics, transcriptomics, proteomics, and metabolomics while preserving spatial architecture, it provides multidimensional, high-resolution molecular maps. These technologies are reshaping our understanding of complex biological systems, particularly in tumor heterogeneity, developmental biology, and disease mechanisms. They will continue to play increasingly important roles in both basic research and clinical translation, offering powerful tools for uncovering biological complexity and advancing disease diagnosis and treatment.
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


