Integrating Spatial Transcriptomics and Metabolomics for Spatial Omics Discovery
Spatial omics integration is transforming our ability to understand biological systems at the cellular and molecular levels. By seamlessly combining spatial transcriptomics and spatial metabolomics, researchers can now map both gene expression and metabolic activity within intact tissues, providing a more comprehensive view of biological processes. This multi-omics spatial analysis approach enables the simultaneous examination of complex molecular interactions, revealing spatially resolved insights into disease mechanisms, developmental biology, and therapeutic responses. The ability to observe how genes and metabolites are spatially distributed within tissues offers unparalleled depth and precision, making spatial omics an indispensable tool for advancing personalized medicine, cancer research, and regenerative therapies. As a result, spatial omics is rapidly becoming a cornerstone of modern biomedical research, empowering scientists to uncover previously hidden patterns and interactions that are critical for understanding health and disease at a deeper, more integrated level.
Challenges in Spatial Multi-Omics Integration: Resolution, Processing, and Platform Obstacles
Integrating spatial multi-omics data is a complex and challenging task, as it involves combining data from different modalities—spatial transcriptomics and spatial metabolomics—each with unique technical requirements. Key challenges that researchers face in this process include:
- Resolution Differences: Spatial transcriptomics and spatial metabolomics typically operate at different resolutions, which can lead to mismatched data when trying to align gene expression with metabolic activity. Achieving a consistent spatial resolution across these modalities is a crucial first step in successful integration.
- Data Processing Inconsistencies: Different data preprocessing methods across platforms can introduce variations that hinder effective integration. Without standardized protocols, it becomes difficult to harmonize the data for accurate multi-omics analysis. Establishing consistent preprocessing workflows across platforms is essential for reducing biases and improving data comparability.
- Cross-Modal Correlation Verification: One of the key obstacles in spatial multi-omics integration is the need for robust statistical models that can differentiate true biological signals from noise. Effective tools and models are required to validate the correlations between gene expression and metabolite distribution, ensuring that observed relationships are biologically meaningful rather than incidental.
- Tissue Heterogeneity: Biological tissues exhibit significant complexity due to the diverse types of cells and varying microenvironments within them. This heterogeneity adds a layer of difficulty in integrating data, as different cell types may show different spatial patterns of gene expression and metabolic activity, which must be accounted for in the analysis.
- Platform-Specific Noise: Each platform used in spatial omics—whether mass spectrometry imaging or sequencing—introduces unique types of noise, such as ion suppression effects or amplification biases. This platform-specific noise can distort the data, making it challenging to merge information from different modalities without careful preprocessing and correction.
Furthermore, the absence of standardized experimental protocols and well-established benchmark datasets complicates the comparison and integration of findings across different studies. Researchers are often left to address these inconsistencies on a case-by-case basis, making multi-omics integration a dynamic yet complex endeavor.
Strategies for Spatial Multi-Omics Integration: Data Coordination and Alignment
Given the challenges outlined above, effective data coordination and spatial alignment are critical for achieving high-quality spatial multi-omics integration. These processes help ensure that the data from different modalities are properly aligned, allowing researchers to draw meaningful biological conclusions from the integrated datasets. Key strategies for overcoming these challenges include:
- Image Registration Techniques: One of the foundational steps in integrating spatial data is multi-modal image alignment. By using morphological markers, fluorescent probes, or histological images, researchers can align data from different modalities on the same tissue section. This ensures that the gene expression data from spatial transcriptomics matches the spatial positions of metabolic activities detected in spatial metabolomics.
- Batch Effect Correction: Technical variations between different experimental batches or platforms can introduce bias into the data. Algorithms like ComBat and Harmony are widely used to mitigate these batch effects, ensuring that the resulting data is free from platform-specific inconsistencies. This step is crucial for ensuring that the data integration reflects biological variability rather than technical noise.
- Cellular Resolution Matching: A common issue in multi-omics integration is the mismatch in resolution between different types of data. Researchers often use interpolation or downsampling techniques to adjust the resolution of one modality to match the other. This helps ensure that data from different platforms are aligned on a consistent scale, enabling seamless integration and analysis.
- Generative Models and Neural Networks: Cutting-edge approaches, such as Variational Autoencoders (VAE) and Generative Adversarial Networks (GAN), are increasingly being used for cross-modal data generation and embedding representation learning. These methods allow for the generation of missing data points and the extraction of meaningful patterns, facilitating more accurate integration of spatial omics datasets.
Accurate alignment in both the spatial and feature dimensions is essential for successful integration. When properly executed, these strategies ensure that the integrated dataset can provide new, meaningful biological insights.
Tools and Platforms for Spatial Multi-Omics Integration: Key Solutions for Analysis
To facilitate spatial multi-omics integration, several tools and platforms have been specifically designed to handle the complexities of multi-modal data. These platforms provide various functionalities that make it easier to align, process, and analyze spatial transcriptomics and spatial metabolomics data. Some of the most widely used tools in the field include:
|
Tool Name |
Main Features |
Applicable Data Types |
|
MISTY |
Multi-modal spatial data integration and feature extraction |
Spatial Transcriptomics (ST) + Metabolomics |
|
SPACE-GM |
Graph model-based multi-omics integration and spatial co-localization analysis |
|
|
CosMx |
High-throughput imaging platform supporting protein and RNA co-detection |
NanoString CosMx SMI |
|
MERSCOPE |
Ultra-multi-channel fluorescence imaging enabling single-cell resolution integration of metabolites and transcriptomics |
Vizgen MERSCOPE |
|
Seurat |
Extended support for spatial omics data integration and visualization |
10x Genomics Visium + Others |
|
Scanpy |
Python package for processing single-cell and spatial omics data |
|
Case Study: Integrating Spatial Multi-Omics to Explore Tumor Microenvironment (TME) in ccRCC
A recent study published in Nature Genetics titled Multi-omic profiling of clear cell renal cell carcinoma identifies metabolic reprogramming associated with disease progression showcases the powerful application of spatial multi-omics integration in tumor microenvironment (TME) research. This study on clear cell renal cell carcinoma (ccRCC) employed both spatial transcriptomics and spatial metabolomics to uncover the spatial relationship between tumor heterogeneity (ITH) and metabolic-immune interactions—key breakthroughs in understanding ccRCC progression.
The study found that, in the immune subtype IM3, which is rich in CD8+ T cells, spatial metabolomics detected significant enrichment of pyrimidine derivatives, such as guanine and hypoxanthine. These metabolites co-localized with areas of CD8+ T cell infiltration, indicating a metabolic environment conducive to T cell activity. Further analysis using spatial transcriptomics revealed upregulation of pyrimidine metabolism pathway genes in these regions, suggesting that the local metabolic microenvironment supports T cell survival or function. Conversely, in the DCCD (dedifferentiated clear cell) subtype IM4 region, spatial metabolomics showed a decrease in long-chain fatty acids and glycerophospholipids, consistent with a phenotype of low lipid droplets. Spatial transcriptomics confirmed widespread inhibition of metabolic pathways such as fatty acid oxidation, and an increase in nutrient transporter gene expression (e.g., SLC family), highlighting altered metabolic pathways in this region.
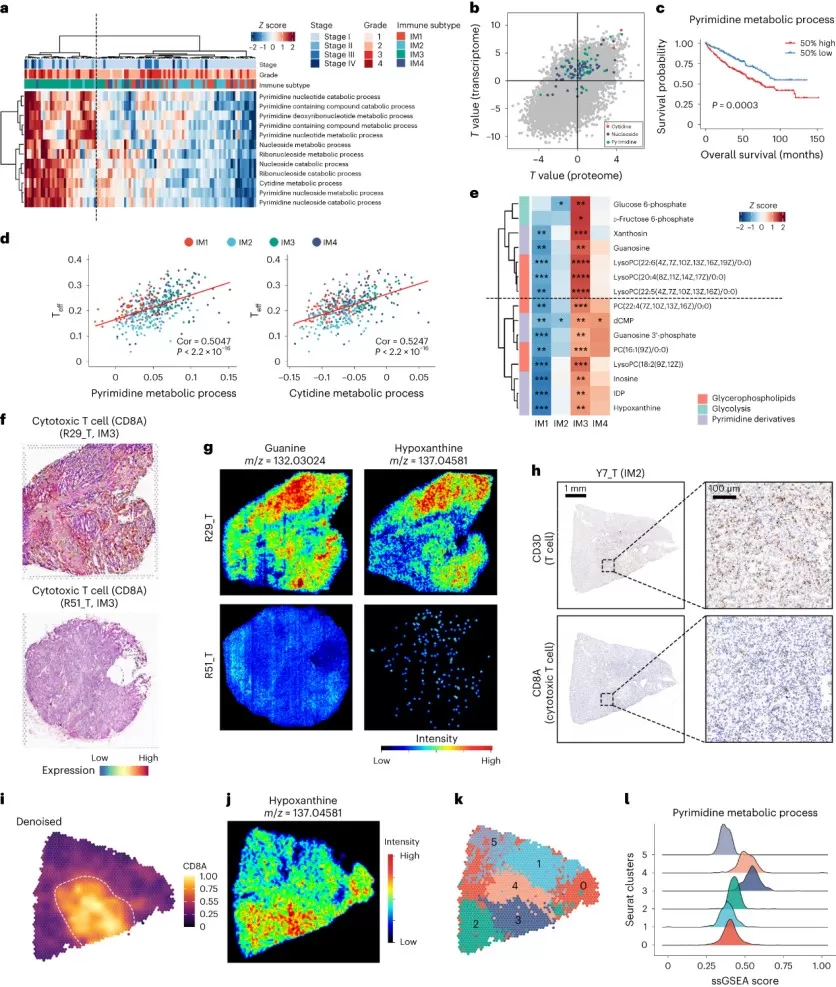
Correlation between immune and metabolic heterogeneity in the TME (Hu et al., 2024)
These findings underscore the value of spatial technologies in revealing deeper insights into tumor biology, as demonstrated by this study:
- Direct visualization of metabolite and gene expression spatial distribution: Linking molecular phenotypes to tissue architecture, providing a clearer understanding of the tissue's biological functions.
- Uncovering metabolic-immune co-localization: This integration enables a more nuanced approach to cancer therapy by highlighting potential targets for combined metabolic and immune-based therapies.
- Identifying tumor heterogeneity: The study revealed the coexistence of DCCD and non-DCCD regions within the tumor, demonstrating that a single biopsy could miss key subclonal populations, underscoring the importance of spatial resolution in capturing tumor complexity.
- Inferring tumor evolution: Through spatial trajectory analysis, the study enhanced our understanding of ccRCC progression, offering insights into how tumors evolve in both metabolic and immune contexts.
In summary, spatial multi-omics technologies were essential for this study, providing spatial resolution that is unattainable through traditional bulk sequencing methods. The integration of spatial transcriptomics and spatial metabolomics allowed for a deeper, more dynamic understanding of how metabolic reprogramming and immune microenvironments interact in ccRCC, ultimately advancing our ability to understand and treat complex tumors.
Best Practices and Common Pitfalls in Spatial Multi-Omics Integration
To ensure the success of spatial multi-omics integration and maximize the value derived from these advanced analytical methods, it is crucial to adhere to best practices at both the experimental design and data analysis stages. In parallel, understanding and avoiding common pitfalls is equally important for achieving reliable, reproducible results.
Best Practices at the Experimental Design Stage:
- Standardize Sample Preprocessing: Consistency is key when preparing samples for multi-omics analysis. Ensure uniformity by using continuous tissue sections or splitting the same tissue section for both spatial transcriptomics and spatial metabolomics. This minimizes variability and ensures that data from different modalities come from the same biological context.
- Incorporate Internal Standards and Negative Controls: Including internal standards (e.g., spike-in controls) and negative controls in your experimental setup helps account for technical errors and background noise. This allows for the accurate detection of biological signals and improves data reliability.
Best Practices at the Data Analysis Stage:
- Use Multiple Spatial Alignment Methods for Cross-Validation: Since spatial omics data often come from different platforms, using various spatial alignment techniques—such as image registration and mutual information—can help cross-validate findings. This ensures that the spatial relationships between different data types are accurately aligned.
- Validate Integration Workflows with Simulated Data or Known Biological Structures: Testing integration pipelines with simulated datasets or previously characterized biological structures allows researchers to assess the robustness and accuracy of their workflows before applying them to experimental data.
- Integrate Functional Enrichment Analysis: To enhance the interpretability of integrated multi-omics data, incorporate functional enrichment analysis to identify pathways and biological processes that are overrepresented. This can reveal biologically relevant patterns in gene and metabolite interactions, further supporting the biological conclusions drawn from the data.
Common Pitfalls:
Despite the best practices, several common pitfalls can compromise the quality of spatial multi-omics integration:
- Relying Solely on Algorithmic Output Without Biological Validation: Algorithms can generate insightful results, but they should never replace biological validation. It is essential to confirm findings through experimental validation or comparison with known biological mechanisms to ensure the results reflect true biological phenomena.
- Ignoring Spatial Autocorrelation: Spatial data is inherently correlated; ignoring spatial autocorrelation can lead to false positives and misinterpretations of the data. Careful statistical methods must be applied to account for spatial dependencies and avoid misleading conclusions.
- Over-Interpreting Correlations as Causal Relationships: While spatial multi-omics integration can reveal correlations between gene expression and metabolite distributions, it is critical not to jump to conclusions about causality without experimental evidence. Correlations do not imply direct cause-and-effect relationships, and further investigation is often necessary.
Future Prospects: Integration and Innovation in Spatial Multi-Omics
The field of spatial multi-omics integration is evolving rapidly, driven by technological innovations and the increasing application of artificial intelligence (AI) in biological research. As these advancements unfold, we can expect several transformative trends in the coming years.
Higher Resolution Technologies:
Emerging techniques, such as single-cell metabolomics imaging and subcellular spatial transcriptomics, are pushing the boundaries of spatial resolution. These technologies allow researchers to probe molecular interactions at the single-cell or even subcellular level, providing a more detailed and accurate picture of biological processes within tissues. This increased resolution will be crucial for uncovering complex biological mechanisms, particularly in heterogeneous tissues like tumors.
Deep AI Applications:
With the growth of AI and machine learning, deep learning-based integration models are expected to play a pivotal role in spatial multi-omics integration. These advanced models will more effectively identify and extract cross-modal signals from vast and complex datasets. AI will also assist in the interpretation of large-scale multi-omics data, enabling researchers to identify subtle biological patterns that might have otherwise been overlooked.
Dynamic and Time-Series Analysis:
There is growing interest in dynamic and time-series analysis of spatial multi-omics data. By studying the temporal changes in gene expression and metabolite distribution within a spatial context, researchers can gain insights into the progression of biological processes, disease development, and therapeutic responses. This type of analysis will be invaluable in understanding disease mechanisms in real-time, including cancer progression, tissue regeneration, and immune response dynamics.
Multi-Modal Data Cloud Platforms:
To manage the increasing complexity of spatial multi-omics data, cloud-based solutions will become essential. Multi-modal data cloud platforms are expected to integrate storage, analysis, and visualization in a seamless, centralized environment. These platforms will improve data accessibility, foster collaborative research, and enable more efficient and scalable analyses. By leveraging cloud infrastructure, researchers will be able to handle larger datasets, perform more complex analyses, and make their findings more widely accessible to the scientific community.
Accelerating Clinical Translation:
The ultimate goal of spatial multi-omics integration is to bring its transformative power into clinical applications. Spatial multi-omics technologies are poised to become invaluable tools in oncology, neuroscience, immunology, and regenerative medicine. By providing deeper insights into disease mechanisms and therapeutic responses, these technologies can facilitate the development of personalized therapies and improve patient outcomes. As clinical translation accelerates, spatial multi-omics could revolutionize diagnostic approaches, enabling more precise disease characterization and tailored treatments.
Reference:
Hu, J., Wang, SG., Hou, Y. et al. Multi-omic profiling of clear cell renal cell carcinoma identifies metabolic reprogramming associated with disease progression. Nat Genet 56, 442–457 (2024). https://doi.org/10.1038/s41588-024-01662-5
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


