Fecal Metabolomics: Decoding Gut Microbiota–Host Interactions Through Microbial Metabolites
The gut microbiome is often referred to as a "hidden organ," exerting profound influence on human health through a complex chemical dialogue with the host. While sequencing technologies have mapped the microbial landscape, fecal metabolomics serves as the essential functional readout, capturing the real-time molecular output of these microbial communities. By analyzing microbiome-derived metabolites, researchers are uncovering novel biomarkers for disease, revealing mechanisms behind the gut-brain axis, and paving the way for personalized nutrition and therapeutics. This blog serves as your comprehensive guide to understanding how fecal metabolomics is revolutionizing our approach to human health and disease.
1. What is Fecal Metabolomics: The Bridge Between Microbiome and Phenotype
Fecal metabolomics is a specialized field within systems biology dedicated to the systematic identification and quantification of the complete set of small-molecule metabolites present in a stool sample. It uniquely bridges the gap between microbial composition and host phenotype by quantifying the functional output—the signaling molecules, nutrients, and byproducts—generated through the dynamic interplay of diet, host cells, and the gut microbiota. This analytical power is anchored in its capacity to provide a definitive functional readout of microbial activity, its utilization of feces as an exceptionally information-rich matrix, and the non-invasive, dynamic monitoring it enables for tracking health and disease interventions over time.
1.1 Fecal Metabolomics as the Definitive Functional Readout
For years, 16S rRNA and metagenomic sequencing have been the gold standards for asking "who is there?" in the gut. However, the presence of a microbe does not guarantee its activity. Fecal metabolomics fills this critical "functional gap" by measuring the end-products of metabolism—the actual molecules produced or modified by the microbiota. This approach treats fecal metabolites as a functional readout, translating the vast genetic data of the microbiome into observable phenotypic traits. It allows researchers to distinguish between a dormant community and an active one, providing the definitive evidence needed to establish causality in host-microbe studies.
1.2 Feces as an Information Rich Matrix for Metabolite Analysis
Feces is often dismissed as waste, but in multi-omics research, it is recognized as a highly information-rich matrix. The fecal metabolome is a complex molecular mixture containing undigested dietary components, host intestinal secretions, and a vast array of microbial metabolites. This includes critical signaling molecules such as short-chain fatty acids (SCFAs), bile acids, and amino acid derivatives, as well as exogenous substances like environmental pollutants or drug residues. By analyzing this diverse chemical soup, scientists can simultaneously assess the impact of diet, the efficiency of microbial fermentation, and the host's metabolic excretion, capturing a complete snapshot of the gut microenvironment.
1.3 Fecal Sampling as a Non-Invasive Tool for Longitudinal Studies
Unlike blood or tissue sampling, fecal collection is non-invasive and patient-friendly, making it an ideal tool for large-scale clinical trials and longitudinal studies. This accessibility allows for dynamic monitoring of how the gut metabolome responds to specific interventions, such as dietary adjustments, probiotic supplementation, or pharmaceutical treatments. Whether tracking the progression of metabolic disease or evaluating a therapeutic response, fecal metabolomics enables the continuous observation of the gut's changing chemical landscape without imposing stress on the subject.
2. Key Metabolite Classes in Fecal Metabolomics
The true value of fecal metabolomics lies in interpreting the specific molecules it reveals. These metabolites are not just chemical endpoints; they are active messengers and regulators within the gut-brain axis and systemic metabolism. By understanding the major classes—Short-Chain Fatty Acids (SCFAs), Bile Acids, and Tryptophan Derivatives—researchers can decipher the functional language of the gut, linking specific microbial activities to host health outcomes and identifying potential biomarkers for disease.
2.1 Short-Chain Fatty Acids (SCFAs): Microbial Metabolites with Systemic Influence
Short-chain fatty acids (SCFAs), primarily acetate, propionate, and butyrate, are produced by gut bacteria through the fermentation of dietary fiber. They are among the most studied and impactful microbiome-derived metabolites.
- Acetate serves as a substrate for other bacteria and peripheral tissues, influencing cholesterol and lipid metabolism.
- Propionate is primarily metabolized in the liver, where it acts as a gluconeogenesis precursor and contributes to satiety signaling.
- Butyrate is the preferred energy source for colonocytes and is crucial for maintaining gut barrier integrity. It also exerts potent anti-inflammatory effects by inhibiting histone deacetylases and modulating immune cell function (Parada Venegas et al., 2019).
Altered SCFA profiles in feces are consistently linked to conditions like inflammatory bowel disease (IBD), obesity, and type 2 diabetes, highlighting their role as key functional mediators in host-microbiota interactions (Silva et al., 2020).
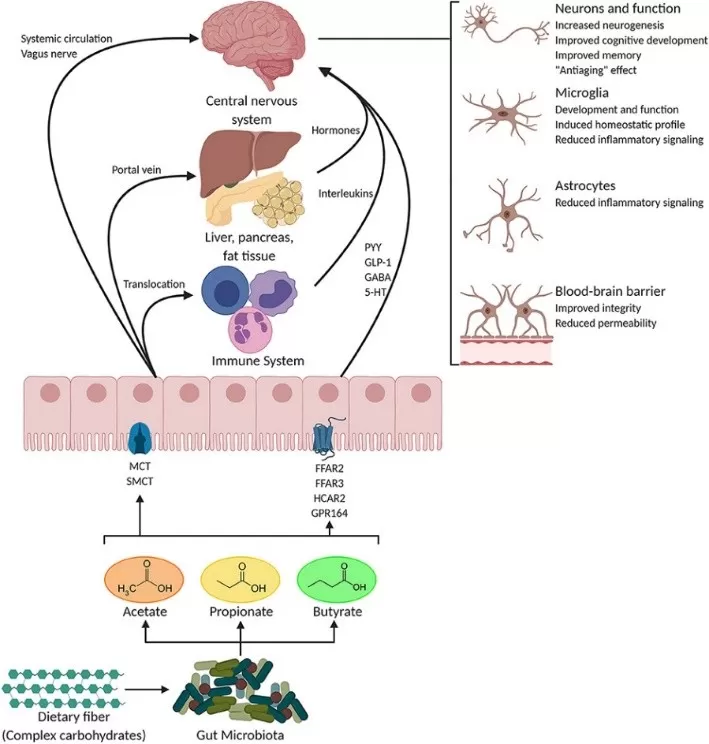
Potential pathways through which SCFAs influence gut-brain communication.
Image reproduced from Silva et al., 2020, Frontiers in endocrinology, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
2.2 Bile Acid Metabolism: A Dialogue Between Liver and Microbiome
Bile acids are classic examples of co-metabolism. Primary bile acids (e.g., cholic acid) are synthesized in the liver, conjugated, and secreted into the intestine. There, gut microbes extensively modify them through deconjugation and dehydroxylation to generate a diverse pool of secondary bile acids (e.g., deoxycholic acid, lithocholic acid). This transformation is critical for efficient lipid digestion and absorption.
More importantly, both primary and secondary bile acids act as signaling molecules, activating receptors such as the farnesoid X receptor (FXR) and the G protein-coupled bile acid receptor 1 (GPBAR1). These receptors regulate host metabolism, immune responses, and energy homeostasis (Jia et al., 2021).
Dysbiosis can disrupt this delicate balance, leading to bile acid pool alterations implicated in metabolic dysfunction-associated steatotic liver disease (MASLD), insulin resistance, and colorectal cancer. Thus, fecal bile acid profiling provides direct insight into this critical liver-gut axis.
2.3 Tryptophan Derivatives: Linking Microbial Metabolism to the Gut-Brain Axis
The metabolism of the essential amino acid tryptophan represents a major intersection point between microbial activity, host physiology, and neurological function. Tryptophan is catabolized through three principal pathways:
- The Microbial Indole Pathway: Gut bacteria convert tryptophan into various indole derivatives (e.g., indole-3-propionic acid, indoxyl sulfate). These metabolites can influence intestinal barrier function and host immunity by activating the aryl hydrocarbon receptor (AhR) (Roager & Licht, 2018).
- The Host Serotonin Pathway: A significant portion of the body's serotonin, a key neurotransmitter, is synthesized in the gut from tryptophan.
- The Host Kynurenine Pathway: This major route produces metabolites that can have neuroactive and immunomodulatory properties.
The balance between these pathways, heavily influenced by the gut microbiota, is crucial. Shifts in tryptophan metabolism and the levels of its fecal derivatives are actively investigated in the context of gut-brain axis disorders, including depression, autism spectrum disorder, and neurodegenerative diseases, offering new avenues for biomarker discovery (Gao et al., 2020).
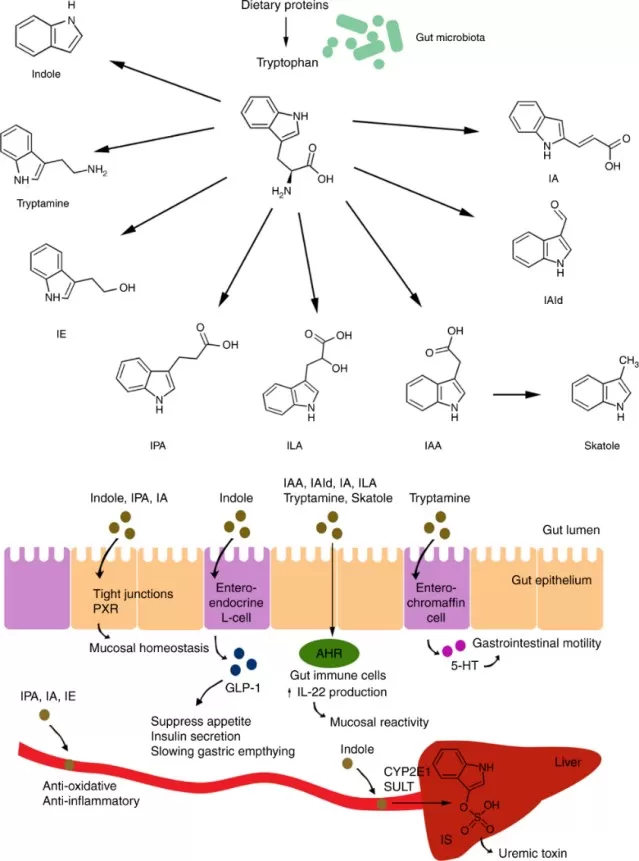
Mechanisms of action of microbial tryptophan catabolites on host physiology.
Image reproduced from Roager & Licht, 2018, Nature communications, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
3. Analytical Platforms and Strategies in Fecal Metabolomics
Transforming a complex fecal sample into reliable, interpretable data requires a carefully designed analytical pipeline. The choice of technology and strategy directly determines the depth, accuracy, and biological relevance of the findings. This chapter outlines the core platforms and methodological approaches in fecal metabolomics, guiding researchers on selecting the right tools to match their specific research objectives, whether for broad-scale biomarker discovery or precise quantitative validation.
3.1 Analytical Platforms: LC-MS/MS, GC-MS, and Their Applications
The two most prevalent platforms in metabolomics are Liquid Chromatography-Mass Spectrometry (LC-MS/MS) and Gas Chromatography-Mass Spectrometry (GC-MS), each with distinct strengths. (Learn more at: LC-MS VS GC-MS: What's the Difference)
i. LC-MS/MS for Broad Metabolite Coverage: Liquid Chromatography-Mass Spectrometry (LC-MS/MS) is the workhorse of modern fecal metabolomics. Its key advantage is exceptional versatility and broad coverage. The liquid chromatography front-end effectively separates a wide range of metabolites with diverse polarities, from hydrophilic amino acids to hydrophobic lipids. Coupled with high-resolution and tandem mass spectrometers, LC-MS/MS enables the detection and tentative identification of thousands of metabolites in a single untargeted analysis. This makes it the ideal platform for discovery-phase studies aiming to map the global metabolic shifts associated with disease or interventions. For targeted analysis, LC-MS/MS provides high sensitivity and specificity for the precise quantification of pre-defined metabolite panels.
ii. GC-MS for Volatile and Derivatizable Compounds: Gas Chromatography-Mass Spectrometry (GC-MS) excels in the analysis of volatile, thermally stable compounds. Its greatest strength in gut research is the highly sensitive and quantitative analysis of short-chain fatty acids (SCFAs), which are central to gut health. While many metabolites require chemical derivatization to become volatile for GC-MS analysis, this process also provides highly reproducible fragmentation patterns and enables comparisons against extensive, standardized spectral libraries. This results in confident metabolite identification. Therefore, GC-MS is often the platform of choice for focused, quantitative studies on SCFAs, and other organic acids.
3.2 Untargeted vs. Targeted Strategies: When to Use Which?
The choice between an untargeted and a targeted strategy is fundamental and depends entirely on the research question.
i. The Untargeted (Discovery) Strategy: Untargeted metabolomics is a hypothesis-generating approach. Its goal is to profile as many metabolites as possible in a sample without prior bias. This is crucial for biomarker discovery, revealing novel metabolic pathways, and understanding global system-wide responses. However, it is inherently semi-quantitative, and a significant portion of detected features may remain unannotated. The workflow involves sophisticated data processing, statistical analysis (e.g., multivariate analysis), and often requires subsequent targeted validation to confirm key findings.
ii. The Targeted (Validation) Strategy: Targeted metabolomics is a hypothesis-driven approach. It focuses on the accurate absolute quantification of a predefined, often smaller, set of metabolites (e.g., a panel of 50 bile acids). This method relies on optimized protocols, authentic standards for each compound, and generates highly accurate, reproducible, and absolute concentration data. It is the gold standard for validating biomarkers from discovery studies, conducting clinical assays, and performing rigorous pathway-focused analyses (e.g., monitoring the tryptophan metabolism pathway). Targeted assays offer higher sensitivity, precision, and are essential for translating discoveries into reliable clinical or mechanistic insights.
iii. Selecting the Right Path: A powerful and common research paradigm is to begin with an untargeted LC-MS/MS screening to identify differentially abundant metabolites, then transition to a customized targeted method (using LC-MS/MS or GC-MS) to validate and precisely quantify these hits in a larger cohort, effectively bridging discovery with robust validation.
4. From Sample to Insight: A Standardized Fecal Metabolomics Workflow
A robust and standardized workflow is the cornerstone of generating reliable, reproducible data in fecal metabolomics. This process transforms a raw biological sample into biologically meaningful insights, with each step—from collection to computational analysis—critically influencing the final results. This chapter outlines the three pillars of this workflow: meticulous sample preparation, rigorous data acquisition and processing, and insightful bioinformatic analysis, providing a blueprint for achieving high-quality data.
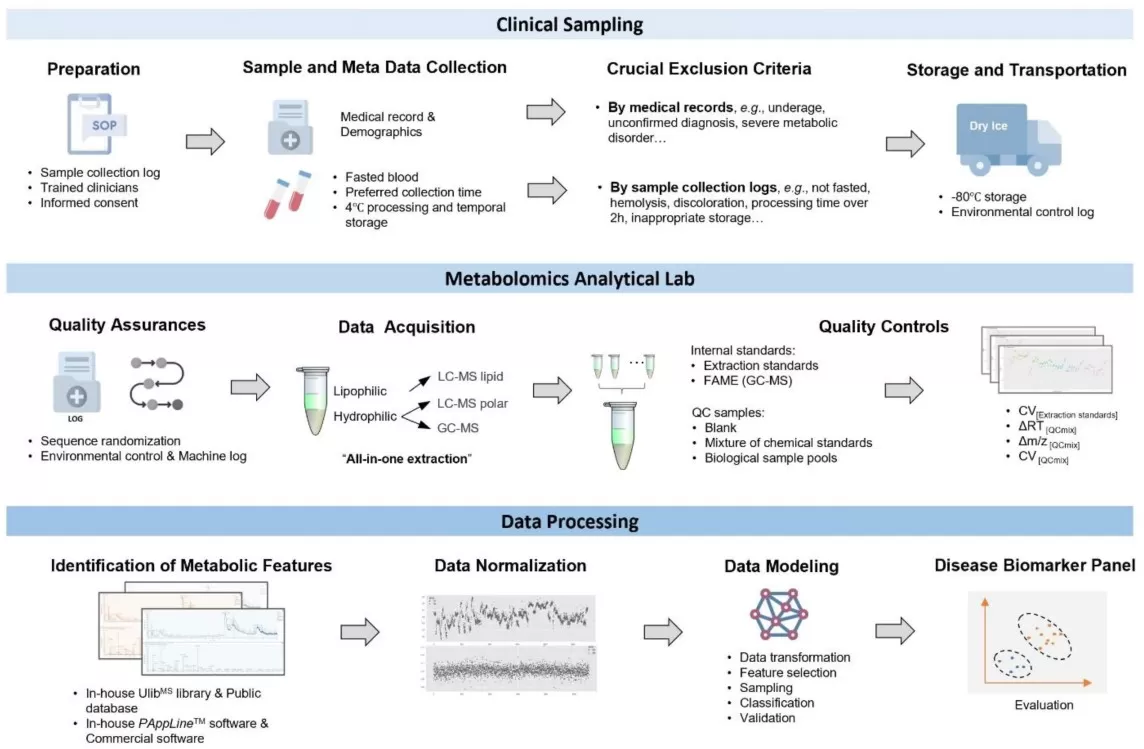
The workflow for a clinical-oriented metabolomics study.
Image reproduced from Shi et al., 2022, Metabolites, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
4.1 Sample Preparation and Pre-analytical Considerations
The journey from sample to data begins long before the instrument analysis. Pre-analytical variables are arguably the most significant source of variation in metabolomics, making standardized protocols paramount.
Fecal material is inherently heterogeneous, both in physical consistency and microbial/metabolite distribution. Therefore, standardized sampling protocols—such as collecting from multiple sites of a stool specimen and homogenizing it in a stabilizing buffer—are essential to obtain a representative aliquot. Immediately following collection, preservation is crucial to halt ongoing microbial activity that would otherwise drastically alter the metabolite profile. The choice between immediate flash-freezing (typically at -80°C) and immersion in a chemical stabilization buffer (e.g., with chaotropes or alcohols) depends on logistics and analytes of interest. Studies have shown that while freezing is excellent for broad-spectrum stability, specific stabilization buffers may better preserve labile metabolites like certain short-chain fatty acids over longer storage or transport periods without constant freezing (De Spiegeleer et al., 2020; Loftfield et al., 2016). Consistent application of one validated method across an entire study cohort is non-negotiable for data integrity.
The extraction step aims to solubilize the maximum range of metabolites while removing proteins and other interfering substances. A common and effective strategy employs a biphasic solvent system, such as methanol/water/chloroform. This approach partitions metabolites based on polarity: hydrophilic compounds (e.g., amino acids, sugars) into the aqueous methanol phase, and hydrophobic lipids into the organic chloroform phase. The choice of solvent ratios and the inclusion of steps like freeze-drying (lyophilization) to remove water and pre-concentrate metabolites can significantly impact extraction efficiency and final detection limits. Optimized and consistent extraction is fundamental for achieving both high coverage in untargeted analysis and accurate quantification in targeted metabolomics.
4.2 Data Acquisition and Processing
Following preparation, samples are analyzed using the chosen platform (e.g., LC-MS/MS). The raw data generated are complex chromatograms and mass spectra, which require sophisticated computational processing to be interpretable. This multi-step pipeline typically includes: peak picking to detect metabolite signals from the background noise; alignment to ensure the same metabolite is matched across all samples despite minor retention time shifts; and normalization to correct for technical variations (e.g., sample weight differences, injection volume inconsistencies). Normalization methods, such as using internal standards, total ion count, or probabilistic quotient normalization, are critical for enabling valid biological comparisons between samples. The output is a cleaned data matrix ready for statistical analysis, with rows representing metabolites (or features) and columns representing samples. (Learn more at: Advanced Techniques in Metabolomics Data Processing)
4.3 Bioinformatics and Statistical Analysis
The processed data matrix is the starting point for biological interpretation. The analysis strategy is tailored to the study design. Unsupervised multivariate statistical methods, like Principal Component Analysis (PCA), are first used to visualize overall data structure, identify outliers, and detect inherent groupings without prior knowledge. Supervised methods, such as Partial Least Squares-Discriminant Analysis (PLS-DA) or Orthogonal PLS-DA (OPLS-DA), are then employed to pinpoint the specific metabolite features that best differentiate predefined groups (e.g., healthy vs. diseased). Significance testing (e.g., t-tests, ANOVA) with appropriate multiple testing correction (like False Discovery Rate) is applied to these features. Finally, to move from a list of significant metabolites to biological understanding, pathway analysis and enrichment analysis tools are used. These map the altered metabolites onto known biochemical pathways (e.g., the tryptophan metabolism pathway or bile acid synthesis), helping to generate mechanistic hypotheses about the underlying biology of the observed condition. (Learn more at: PCA vs PLS-DA vs OPLS-DA)
5. Multi-omics Integration: Fecal Metabolomics + Microbiome
While powerful on its own, the true transformative potential of fecal metabolomics is unlocked through integration with microbiome sequencing data, a paradigm known as multi-omics integration. This approach moves beyond parallel observation to establish functional connections, effectively answering the critical questions of “who is doing what?” By correlating microbial taxa (from 16S rRNA or metagenomic sequencing) with metabolite levels (from fecal metabolomics), researchers can construct interaction networks that pinpoint which microbes are likely producers or modifiers of key bioactive molecules. For instance, integrative studies have revealed how specific bacterial genera, such as Faecalibacterium and Bacteroides, are significantly correlated with shifts in amino acid and bile acid metabolism in gastrointestinal diseases, thereby providing testable hypotheses about the microbial drivers of metabolic dysregulation (Wang et al., 2023). Ultimately, this Microbiome + Metabolome synergistic strategy is indispensable for elucidating complex host-microbiota interactions in conditions like IBD, obesity, and neurological disorders, bridging the gap from microbial community structure to functional pathophysiology and identifying novel therapeutic targets.
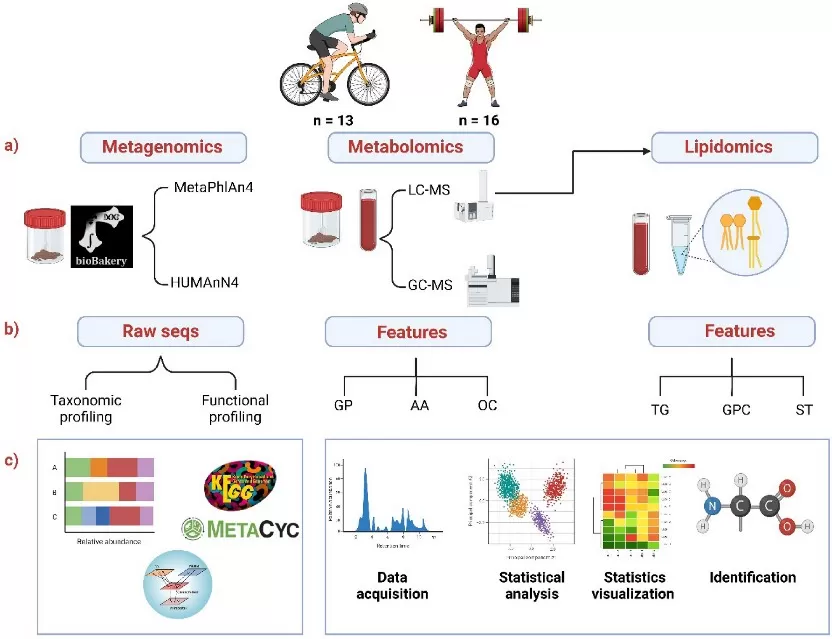
Overview of the integrative omics workflow for studying the gut microbiota and host metabolism in athletes
Image reproduced from Aya et al., 2025, Scientific reports, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
6. Frontier Applications of Fecal Metabolomics in Health and Disease Research
Fecal metabolomics has moved from a descriptive profiling tool to a genuinely translational “metabolite analysis” strategy that links the gut microbiome to actionable biology. Because stool contains both host compounds and microbiome-derived metabolites, it can reveal disease mechanisms, enable biomarker discovery, and provide an objective readout of intervention response—often earlier than clinical endpoints. Below are four of the most active application areas where LC-MS metabolomics (and complementary platforms) is already shaping study design and interpretation.
6.1 Metabolic Diseases: Mechanism Insights and Biomarker Discovery in Type 2 Diabetes
In metabolic disease research, fecal metabolomics is increasingly used to pinpoint which microbial functions—and which downstream metabolites—track with insulin resistance, dyslipidemia, and glycemic control. A common pattern is that shifts in lipid-related molecules and bile acid pools co-occur with microbiome remodeling, suggesting a metabolic “language” through which the gut communicates with host tissues. In a clinical very-low-calorie restriction study in people with type 2 diabetes, investigators combined fecal microbiota profiling with untargeted metabolomics in plasma and feces and observed significant improvements in glucose and lipid parameters alongside measurable changes in fecal metabolites, including decreases in a bile acid–related metabolite (glycholic acid) and a lipid metabolite (LysoPC (18:1)), highlighting how fecal metabolomics can capture intervention-linked pathway shifts rather than only taxonomic changes in the microbiome (Gong et al., 2024).
 and the significant changes of fecal metabolite_1770082111_WNo_515d459.webp)
OPLS-DA of fecal metabolites before and after caloric restriction (C) and the significant changes of fecal metabolites before and after 9-day very-low-calorie restriction (D).
Image reproduced from Gong et al., 2024, Frontiers in endocrinology, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
6.2 Gastrointestinal Disease and Oncology: Mapping Microbiome–Metabolite Drivers in Colorectal Cancer Models
For gastrointestinal disorders—especially inflammation-associated tumorigenesis—fecal metabolomics provides a practical way to connect microbial community changes to host pathways such as epithelial barrier integrity, immune signaling, and pro-tumor vs anti-tumor metabolite production. This is particularly valuable when interventions (diet, antibiotics, probiotics, FMT, or drugs) reshape the gut ecosystem and the metabolic environment simultaneously. In an open-access study of colitis-associated colorectal cancer in mice, researchers integrated gut microbial profiling with metabolomics and showed that fecal microbiota transplantation (FMT) from different human donor groups led to divergent tumor phenotypes; importantly, they also reported specific metabolite–microbe correlations (including links between bacterial taxa and bile-acid–related and lipid-like metabolites), illustrating how fecal metabolomics can help move from “association” toward plausible mechanistic chains in CRC-related microbiome research (Song et al., 2024).
6.3 Gut–Brain Axis: Tryptophan Metabolism Pathway Signals in Neuropsychiatric Research
The gut-brain axis is one of the hottest areas for fecal metabolomics because neuroactive signaling often depends on metabolites produced or transformed in the intestine, including short-chain fatty acids, indoles, and intermediates in the tryptophan metabolism pathway (serotonin-related routes, kynurenine branch, and microbial indole derivatives). Fecal metabolomics offers a direct way to test whether microbiome alterations are accompanied by a corresponding shift in neuroactive chemistry, which is crucial for strengthening biological plausibility in psychiatric and neurological studies. A notable open-access Microbiome paper integrated microbiome profiling with targeted tryptophan metabolomics in adolescent depression and linked specific microbial changes with altered tryptophan-derived neurotransmitter balance; in mouse experiments, transplanting microbiota from healthy adolescents improved depressive-like behaviors and shifted the tryptophan pathway toward higher serotonin (5-HT) and reduced kynurenine-pathway toxic metabolites, providing a compelling example of how metabolite-focused readouts can anchor gut–brain hypotheses in measurable chemistry (Zhou et al., 2023).
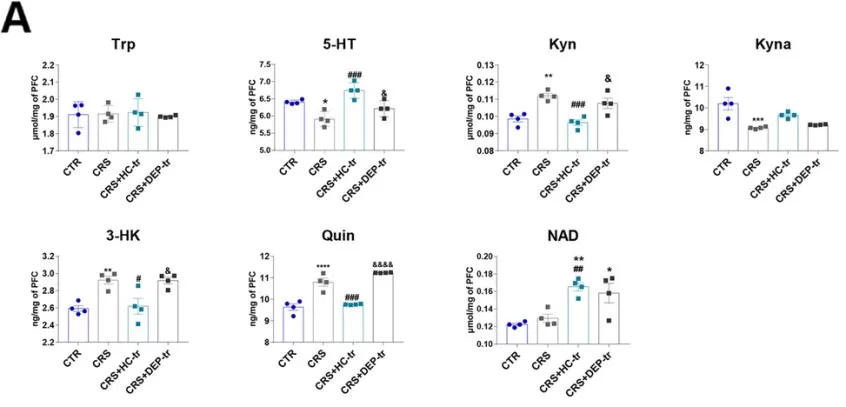
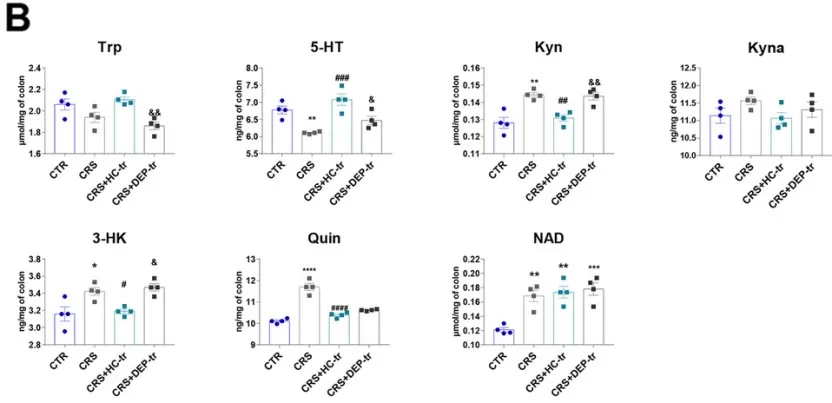
FMT from healthy volunteers ameliorated the neurotransmitter perturbation by the Trp-Kyn pathway by affecting the rate-limiting enzymes in both the brain and colon. A. Levels of Trp-Kyn metabolic pathway-derived neurotransmitters in the mice prefrontal cortexs. B. Levels of Trp-Kyn metabolic pathway-derived neurotransmitters in the mice colons.
Image reproduced from Zhou et al., 2023, Microbiome, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
6.4 Nutrition and Lifestyle Interventions: Objective Readouts of Diet, Fasting, and Metabolic Remodeling
Diet and lifestyle trials often struggle with adherence measurement and with translating “what people did” into “what biology changed.” Fecal metabolomics helps bridge that gap by providing an objective signature of gut metabolic output, including shifts in lipid-like molecules, xenobiotic metabolites, and pathways connected to bile acid metabolism and microbial fermentation (including short-chain fatty acids in many designs, depending on the platform). A 2024 open-access study used fecal untargeted metabolomics (UPLC-HRMS) in mice undergoing intermittent fasting vs normal feeding and applied interpretable machine learning to identify discriminating fecal metabolites as candidate biomarkers of fasting patterns, demonstrating how stool-based metabolite signatures can function as compliance- and response-linked readouts in nutrition studies (Hu et al., 2024).
7. Choosing the Right Partner for Your Fecal Metabolomics Research
The journey from a raw fecal sample to groundbreaking biological insight is complex, requiring precision at every step. The choice of a collaborative partner is therefore not merely a logistical decision, but a strategic one that can define the success and impact of your research. An ideal partner combines cutting-edge technology with deep scientific expertise and a commitment to translating your research questions into robust, interpretable data. As a global leader in metabolomics solutions, MetwareBio is uniquely positioned to be that partner, offering an integrated suite of services designed to unlock the full potential of your fecal metabolomics studies.
Our Comprehensive Service Portfolio
We provide end-to-end solutions tailored to every stage of your research, from initial discovery to targeted validation.
- Untargeted Metabolomics (Discovery Phase): Our broad-spectrum untargeted metabolomics service, powered by advanced LC-MS/MS platforms, is your premier tool for hypothesis generation. We detect and relatively quantify thousands of metabolites in a single run, enabling the unbiased discovery of novel biomarkers and metabolic pathways altered in disease states or in response to interventions. This service is the perfect starting point for mapping the complex gut microbiome and host-microbiota interactions.
- Targeted Metabolomics (Validation & Quantification Phase): To build rigorous, publication-ready data, we offer precise, absolute quantification of key metabolite panels. Our specialized assays are directly relevant to the critical findings discussed in this blog:
i. Bile Acid Targeted Metabolomics: Accurately profiles the full spectrum of primary and secondary bile acids, providing direct insight into the gut-liver axis and its role in metabolic diseases.
ii. SCFA Targeted Metabolomics: Precisely quantifies short-chain fatty acids like acetate, propionate, and butyrate using optimized GC-MS methods, delivering essential data on microbial fermentation and immune modulation.
iii. Tryptophan Pathway Targeted Metabolomics: Focuses on the complete tryptophan metabolism pathway, quantifying key intermediates to elucidate its pivotal role in the gut-brain axis and immune regulation.
- Integrated Microbiome & Metabolomics Analysis (Mechanistic Insight Phase): To answer the pivotal question of “which microbes are doing what?”, we offer combined 16S rRNA gene sequencing or shotgun metagenomics with fecal metabolomics. Our expert bioinformaticians perform sophisticated multi-omics integration analyses, including correlation network construction and pathway mapping. This powerful service moves beyond association to reveal potential causal relationships between specific microbial taxa and key metabolic shifts, providing the mechanistic insights needed for high-impact publications.
Why Partner with MetwareBio?
- Advanced Technology Platform: Our core laboratory is equipped with state-of-the-art high-resolution and tandem mass spectrometers (including QTRAP® and QTOF® platforms), coupled with ultra-performance liquid chromatography (UPLC) and gas chromatography (GC) systems. This ensures the highest sensitivity, resolution, and reproducibility for both untargeted and targeted analyses.
- Deep Scientific Expertise & Rich Experience: Our team of PhD-level scientists and bioinformaticians possesses extensive experience in designing and executing fecal metabolomics projects across diverse fields, from nutritional intervention assessment to neurological disease research. We understand the unique challenges of fecal sample preparation and data interpretation.
- Professional, End-to-End Support: We partner with you from study design to biological insight. Our service includes professional consultation on sample collection, robust data processing using proprietary and industry-standard software, and comprehensive bioinformatics analysis—including pathway enrichment and multi-omics integration—to transform raw data into clear, actionable biological stories.
- Customized and Flexible Solutions: We recognize that every research project is unique. Beyond our standard panels, we offer custom method development to quantify specific metabolites of interest, ensuring your precise research objectives are met.
Contact our scientific team today to discuss your project, request a quote, or learn how our tailored metabolomics services can accelerate your path to discovery.
Refenrence
1. Parada Venegas, D., De la Fuente, M. K., Landskron, G., González, M. J., Quera, R., Dijkstra, G., Harmsen, H. J. M., Faber, K. N., & Hermoso, M. A. (2019). Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Frontiers in immunology, 10, 277. https://doi.org/10.3389/fimmu.2019.00277
2. Silva, Y. P., Bernardi, A., & Frozza, R. L. (2020). The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Frontiers in endocrinology, 11, 25. https://doi.org/10.3389/fendo.2020.00025
3. Jia, W., Xie, G., & Jia, W. (2018). Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nature reviews. Gastroenterology & hepatology, 15(2), 111–128. https://doi.org/10.1038/nrgastro.2017.119
4. Roager, H. M., & Licht, T. R. (2018). Microbial tryptophan catabolites in health and disease. Nature communications, 9(1), 3294. https://doi.org/10.1038/s41467-018-05470-4
5. Gao, K., Mu, C. L., Farzi, A., & Zhu, W. Y. (2020). Tryptophan Metabolism: A Link Between the Gut Microbiota and Brain. Advances in nutrition (Bethesda, Md.), 11(3), 709–723. https://doi.org/10.1093/advances/nmz127
6. Loftfield, E., Vogtmann, E., Sampson, J. N., Moore, S. C., Nelson, H., Knight, R., Chia, N., & Sinha, R. (2016). Comparison of Collection Methods for Fecal Samples for Discovery Metabolomics in Epidemiologic Studies. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology, 25(11), 1483–1490. https://doi.org/10.1158/1055-9965.EPI-16-0409
7. De Spiegeleer, M., De Graeve, M., Huysman, S., Vanderbeke, A., Van Meulebroek, L., & Vanhaecke, L. (2020). Impact of storage conditions on the human stool metabolome and lipidome: Preserving the most accurate fingerprint. Analytica chimica acta, 1108, 79–88. https://doi.org/10.1016/j.aca.2020.02.046
8. Wang, C., Yu, X., Lin, H., Wang, G., Liu, J., Gao, C., Qi, M., Wang, D., & Wang, F. (2023). Integrating microbiome and metabolome revealed microbe-metabolism interactions in the stomach of patients with different severity of peptic ulcer disease. Frontiers in immunology, 14, 1134369. https://doi.org/10.3389/fimmu.2023.1134369
9. Shi, Z., Li, H., Zhang, W., Chen, Y., Zeng, C., Kang, X., Xu, X., Xia, Z., Qing, B., Yuan, Y., Song, G., Caldana, C., Hu, J., Willmitzer, L., & Li, Y. (2022). A Comprehensive Mass Spectrometry-Based Workflow for Clinical Metabolomics Cohort Studies. Metabolites, 12(12), 1168. https://doi.org/10.3390/metabo12121168
10. Aya, V., Pardo-Rodriguez, D., Vega, L. C., Cala, M. P., & Ramírez, J. D. (2025). Integrating metagenomics and metabolomics to study the gut microbiome and host relationships in sports across different energy systems. Scientific reports, 15(1), 15356. https://doi.org/10.1038/s41598-025-98973-2
11. Gong, T., Di, H., Hu, Y., Xu, S., Chen, J., Chen, G., Wei, X., & Liu, C. (2024). Gut microbiota and metabolites exhibit different profiles after very-low-caloric restriction in patients with type 2 diabetes. Frontiers in endocrinology, 14, 1289571. https://doi.org/10.3389/fendo.2023.1289571
12. Song, Q., Gao, Y., Liu, K., Tang, Y., Man, Y., & Wu, H. (2024). Gut microbial and metabolomics profiles reveal the potential mechanism of fecal microbiota transplantation in modulating the progression of colitis-associated colorectal cancer in mice. Journal of translational medicine, 22(1), 1028. https://doi.org/10.1186/s12967-024-05786-4
13. Zhou, M., Fan, Y., Xu, L., Yu, Z., Wang, S., Xu, H., Zhang, J., Zhang, L., Liu, W., Wu, L., Yu, J., Yao, H., Wang, J., & Gao, R. (2023). Microbiome and tryptophan metabolomics analysis in adolescent depression: roles of the gut microbiota in the regulation of tryptophan-derived neurotransmitters and behaviors in human and mice. Microbiome, 11(1), 145. https://doi.org/10.1186/s40168-023-01589-9
14. Hu, X., Xu, Q., Ma, X., Li, L., Wu, Y., & Sun, F. (2024). An interpretable machine learning model for precise prediction of biomarkers for intermittent fasting pattern. Nutrition & metabolism, 21(1), 106. https://doi.org/10.1186/s12986-024-00876-y
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


