Phosphopeptide Enrichment for Phosphoproteomics: TiO2 vs IMAC vs Antibody-Based Methods
Phosphoproteomics enables high-resolution, site-specific mapping of protein phosphorylation, a central regulatory mechanism that drives cell signaling and disease biology. Yet, because phosphopeptides are typically rare in complex digests and span an extreme dynamic range, robust enrichment is often the key step that determines whether phosphorylation events can be confidently detected and quantified. In this article, we provide a practical, method-focused guide to phosphopeptide enrichment—explaining the core principles, performance trade-offs, and selection logic behind TiO2, IMAC, and antibody-based approaches—so you can choose the right workflow for your sample type, research question, and throughput requirements.
Why Phosphopeptide Enrichment is Essential for Phosphoproteomics
Phosphoproteomics refers to the large-scale identification and quantification of phosphorylation sites (commonly on serine, threonine, and tyrosine residues) using mass spectrometry. Because phosphorylation is reversible, dynamic, and tightly coupled to signaling cascades, phosphoproteomics has become a foundational tool for studying mechanisms in cancer, cardiovascular disease, and neurodegenerative disorders, as well as for dissecting kinase-driven pathways and treatment response.
In real-world LC–MS/MS workflows, however, phosphorylation is challenging to measure directly from unfractionated peptide mixtures. After proteolytic digestion, phosphopeptides are often present at very low abundance—frequently below 1% of the total peptide population—while non-phosphorylated peptides dominate the ion current. At the same time, phosphorylation spans a wide dynamic range: a small set of highly abundant phosphopeptides may be detectable, whereas many biologically meaningful phosphorylation events remain hidden below the detection limit. This creates a practical bottleneck where instrument time alone cannot compensate for poor detectability, especially in complex matrices such as whole-cell lysates, tissues, and clinical specimens.
This is precisely why phosphopeptide enrichment has become a core, near-mandatory step in modern phosphoproteomics. By selectively capturing phosphopeptides and reducing background interference from non-phosphorylated peptides, enrichment increases effective sensitivity, improves MS/MS sampling of phosphorylation events, and expands phosphorylation-site coverage—often determining whether you obtain a shallow, biased snapshot or a deep, interpretable phosphoproteome [1]. Importantly, the enrichment strategy you choose also shapes what you see: different chemistries can favor mono- vs. multi-phosphorylated peptides, acidic vs. basic peptides, or specific phosphorylation types (e.g., pY). Therefore, selecting an enrichment method is not just a “sample prep detail”—it is an upstream decision that directly impacts biological conclusions, pathway interpretation, and downstream validation planning.
Core Phosphopeptide Enrichment Methods in Phosphoproteomics
Phosphopeptide enrichment is fundamentally driven by the chemistry of the phosphate group: it carries strong negative charge and exhibits distinct coordination behavior and polarity compared with non-phosphorylated peptides. Modern enrichment strategies exploit these properties by creating selective interactions between phosphopeptides and an affinity medium—either through metal-oxide Lewis acid sites, immobilized multivalent metal ions, or phosphorylation-specific antibodies. Accordingly, the three mainstream approaches used in phosphoproteomics are TiO2-based metal oxide affinity chromatography (TiO2/MOAC), immobilized metal affinity chromatography (IMAC; typically Fe3+, Ti4+, or Zr4+), and antibody-based immunoaffinity enrichment (such as anti-pY or anti-pS/pT capture). Although all three approaches aim to increase phosphopeptide signal and reduce sample complexity, they differ in binding mechanisms and peptide-class preferences.
TiO2 Phosphopeptide Enrichment (Metal Oxide Affinity Chromatography)
TiO2-based phosphopeptide enrichment is a classic implementation of metal oxide affinity chromatography (MOAC). In acidic loading conditions (commonly pH < 3), phosphate groups form strong multidentate coordination interactions with Lewis acid sites on the TiO2 surface, enabling selective retention of phosphopeptides while many non-phosphorylated peptides remain unbound. Elution is typically achieved under basic conditions (often pH > 9), where the interaction is disrupted and phosphopeptides are released. In practice, TiO2 is widely adopted because it is cost-effective, scalable, and compatible with both manual and automated workflows, making it a frequent first choice for global discovery phosphoproteomics in complex samples.
A key practical consideration for TiO2 is controlling non-specific adsorption, especially from acidic non-phosphorylated peptides (rich in Asp/Glu) that can partially mimic phosphate-mediated interactions. To mitigate this, protocols commonly add “exclusion agents” such as lactic acid or 2,5-dihydroxybenzoic acid (DHB) to the loading buffer to competitively suppress non-specific binding without stripping true phosphopeptides [2]. Material format also matters: TiO2 nanoparticles (e.g., Titansphere) can improve performance by increasing surface area and binding capacity, which can be especially beneficial for low-input samples or high-complexity digests where binding sites become limiting.
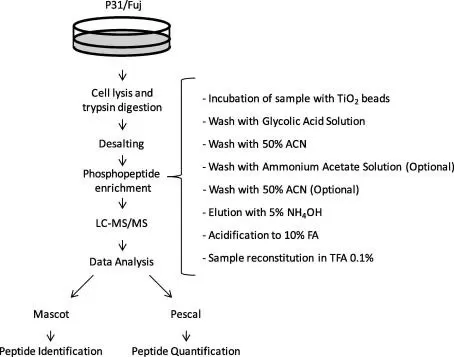
Figure 1. Workflow of TiO2-based phosphopeptide enrichment, including sample lysis, proteolytic digestion, phosphopeptide enrichment, and elution steps.
IMAC Phosphopeptide Enrichment (Immobilized Metal Affinity Chromatography)
IMAC enriches phosphopeptides using immobilized multivalent metal ions—most commonly Fe3+, Ti4+, or Zr4+—that are chelated to a solid support through ligands such as iminodiacetic acid (IDA) or nitrilotriacetic acid (NTA). Phosphopeptides bind through a combination of electrostatic attraction and coordination chemistry between the phosphate group and the metal center. Compared with TiO2, IMAC offers a distinctive advantage: selectivity can be “tuned” by choosing different metal ions and conditions, allowing researchers to bias capture toward particular phosphorylation patterns and peptide chemistries. Elution is commonly performed by shifting pH (weakening coordination) or using competitive reagents (e.g., phosphate-containing buffers) that displace bound phosphopeptides.
From a workflow-design perspective, IMAC is frequently positioned as complementary to TiO2 rather than a direct replacement. Different metal ions can exhibit different affinities for mono- versus multi-phosphorylated peptides; for example, Ti4+-IMAC is often described as having stronger affinity for multi-phosphorylated peptides, whereas Fe3+-IMAC is commonly used for mono-phosphorylated peptides [10]. This tunability makes IMAC especially useful when the project goal is to expand coverage across peptide classes or when TiO2 results show systematic bias. In addition, IMAC can integrate cleanly with quantitative phosphoproteomics workflows, and it is often selected for studies where enrichment performance must remain stable across many samples and batches.
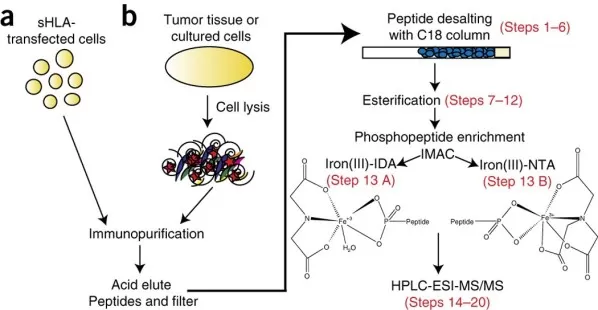
Figure 2: Diagram illustrating the Fe3+-IMAC phosphopeptide enrichment protocol that uses either nitrilotriacetic acid (NTA) or iminodiacetic acid (IDA) columns [3].
Antibody-Based Phosphopeptide Enrichment (Immunoaffinity)
Antibody-based immunoaffinity enrichment captures phosphopeptides through antigen–antibody recognition rather than coordination chemistry. Phosphorylation-specific antibodies—commonly anti-phosphotyrosine (anti-pY) or motif-/context-aware antibodies for phosphoserine/phosphothreonine (pS/pT)—bind phosphopeptides carrying the corresponding epitope, after which immune complexes are isolated using magnetic beads or agarose resins in an immunoprecipitation (IP)-like workflow. Because binding is driven by epitope recognition, immunoaffinity enrichment can deliver exceptionally high specificity and is particularly valuable when the biology is concentrated in a narrow signaling axis (for example, tyrosine kinase pathways) or when the goal is to validate phosphorylation changes in low-abundance targets [9].
The strength of antibody enrichment—its specificity—also defines its constraints. Coverage is inherently limited by antibody panels and epitope compatibility, and performance can depend on sequence context around the phosphorylation site. In other words, immunoaffinity is not designed to be the most comprehensive “global” enrichment approach; instead, it is best viewed as a precision tool for targeted phosphoproteomics questions where purity and interpretability outweigh maximum site count. Commercial multi-antibody mixtures (such as PTMScan-style formats) can broaden capture across motifs and pathways, improving practical coverage while preserving the core advantage of immunoaffinity: a high-confidence enrichment profile that is often difficult to achieve with purely chemistry-driven methods.
How Enrichment Methods Shape Phosphoproteomics Results
Although TiO2, IMAC, and antibody-based immunoaffinity enrichment all aim to enrich phosphopeptides, boost phosphopeptide signal, and reduce sample complexity prior to LC–MS/MS, they do so through distinct capture chemistries. As a result, these methods can favor different phosphopeptide subpopulations and lead to meaningful differences in downstream phosphoproteomics outcomes. In the following sections, we compare how enrichment choice translates into key data-level consequences—most notably specificity, recovery, and overall workflow suitability across different sample types and study goals.
3.1 Phosphopeptide Enrichment Specificity
Studies have shown that TiO2 and IMAC both exhibit high specificity (over 90%), with strong complementarity: TiO2 is more effective for multi-phosphorylated peptides (>2 sites, recovery rate 80-90%), while IMAC (e.g., Fe3+-IMAC) is better for basic, mono-phosphorylated peptides (specificity 85-95%). Antibody methods, however, provide the highest specificity (>95%) but are limited to specific types of phosphorylation, such as anti-pY, which covers tyrosine phosphorylation at 95% but overlooks serine/threonine sites (90%) [7]. A 2020 study showed that TiO2 exhibited less than 5% non-specific binding in complex samples (using DHB exclusion agents), IMAC had 10-15%, and antibody methods showed less than 1% [8].
Furthermore, specificity is significantly influenced by exclusion agents: The lactic acid and glutamic acid methods demonstrate high enrichment specificity, with averages of 88.65% and 86.39%, respectively. In contrast, methods using glycolic acid or DHB exhibit significantly lower specificity, averaging only 52.90% and 41.54% [6]. The choice of metal ions in IMAC is crucial, with Zr4+-IMAC providing higher specificity for pY sites but lower recovery rates. Batch-to-batch variation in antibody methods can be minimized through standardized protocols (e.g., polyclonal antibody mixtures). In HeLa cell lysate experiments, TiO2 identified over 10,000 phosphorylation sites, IMAC identified about 8,000, and antibody methods identified only a few hundred, but with an impressive purity of 98% [4].
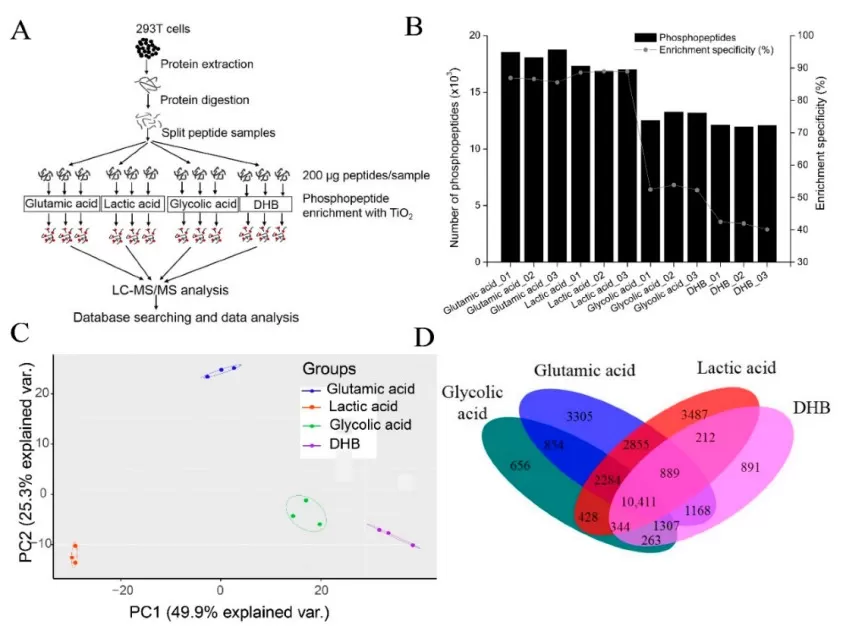
Figure 3. Comparison of four TiO2-based phosphopeptide enrichment methods using different non-phosphopeptide excluders [6].
3.2 Phosphopeptide Enrichment Recovery
Recovery is strongly influenced by input amount, buffer composition, and upstream sample preparation. Under typical conditions, TiO2 enrichment often delivers a recovery of ~70–85% (for nanogram-level inputs), which is generally higher than ~60–75% reported for IMAC; antibody-based immunoaffinity enrichment is usually lower (~40–60%), but it can be particularly valuable when targeting very low-abundance phosphorylation events. A 2017 evaluation reported that TiO2 achieved ~80% recovery on an automated platform (e.g., AssayMAP Bravo), whereas manually performed IMAC reached ~65% [9]. Method sensitivity to experimental variation is also distinct: for example, pH drift can reduce TiO2 recovery by ~20%, while IMAC performance can be compromised by metal contamination. Importantly, sequential workflows such as SIMAC (IMAC followed by TiO2) can push overall recovery to >90% by capturing complementary phosphopeptide subsets across steps [5].
Beyond the enrichment step itself, several upstream variables can reshape the peptide pool and indirectly affect phosphopeptide capture. Trypsin digestion time influences peptide-length distribution; overly long peptides can reduce TiO2 enrichment efficiency due to less favorable binding and/or steric effects during adsorption. Storage conditions (e.g., −80 °C versus room temperature) may have limited short-term impact, but repeated freeze–thaw cycles can accelerate degradation and reduce the integrity of phosphorylation sites. Buffer optimization is therefore not optional: for instance, adding 0.5–1% TFA in TiO2 workflows has been reported to improve recovery by ~10–15% [10]. In multicenter settings, a major source of variability is operator-to-operator differences; standardized, robotic platforms can reduce the coefficient of variation (CV) to <5%. For clinical matrices, matrix effects can depress IMAC recovery to ~50%, whereas TiO2 can maintain around ~70% when paired with appropriate exclusion reagents [6].
3.3 Applicable Scenarios of Different Phosphopeptide Enrichment Methods
TiO2 phosphopeptide enrichment is well suited for high-throughput, discovery-oriented phosphoproteomics, for example large-scale cancer phosphoproteome mapping where studies often report identification of >10,000 phosphorylation sites; IMAC-based phosphopeptide enrichment is frequently used in workflows tied to protein complex or interaction-focused studies and integrates smoothly with downstream quantitative strategies (e.g., iTRAQ); antibody-based immunoaffinity enrichment is most appropriate for targeted phosphoproteomics, such as pathway-centric interrogation of signaling nodes (for instance, EGFR-related phosphorylation events). In highly complex samples, combining TiO2 and IMAC phosphopeptide enrichment can be particularly powerful because the two chemistries tend to capture overlapping but non-identical phosphopeptide populations: TiO2 preferentially retains more acidic and multi-phosphorylated peptides, whereas IMAC often enriches more basic and mono-phosphorylated peptides, thereby improving overall phosphosite coverage. For clinical matrices (e.g., plasma), antibody-based enrichment can reduce matrix-related interference by selectively pulling down the desired phosphopeptide class, although the approach is typically associated with higher reagent cost. A multi-center study further reported that, under standardized protocols, the coefficient of variation (CV) was <10% for TiO2, <15% for IMAC, and <5% for antibody enrichment [7].
A common expanded combination strategy is SIMAC, where IMAC is first applied to enrich mono-phosphorylated peptides, followed by TiO2 enrichment of the IMAC flow-through to capture additional phosphopeptides and increase overall coverage by approximately 30% [1]. For phosphotyrosine (pY)-focused studies, anti-pY immunoaffinity enrichment coupled with TiO2 is often used to balance high selectivity with broader phosphopeptide capture. In addition, automated platforms such as EasyPhos enable TiO2-based processing in 96-well formats, supporting throughput of hundreds of samples per day in large-scale studies [8]. These workflows can also translate to non-mammalian systems (including plants and bacteria), where IMAC-based enrichment is often considered more stable across divergent phosphorylation contexts. Looking ahead, emerging nanocomposite materials—such as magnetic TiO2—are expected to simplify handling and reduce sample loss during phosphopeptide enrichment workflows [10].
TiO2 vs IMAC vs Antibody Enrichment in Phosphoproteomics: A Side-by-Side Comparison
Selecting a phosphopeptide enrichment method is one of the most consequential design choices in phosphoproteomics because the three mainstream approaches—TiO2 phosphopeptide enrichment, IMAC phosphopeptide enrichment, and antibody-based immunoaffinity enrichment—are built on fundamentally different capture principles (metal-oxide Lewis acid interactions, chelated multivalent metal-ion coordination, and phosphorylation-epitope recognition, respectively). These mechanistic differences do not only determine which phosphopeptides are preferentially retained; they also translate into clearly different experimental outcomes, including the specificity and recovery trends discussed in the previous chapter, as well as differences in robustness across sample matrices and project types. As a result, each method has a distinct “best-fit” application range—from high-throughput global phosphoproteome profiling, to complementary deep-coverage workflows, to highly targeted pathway or clinical validation studies. To make these trade-offs easy to evaluate at a glance, the table below consolidates the core principles and the most decision-relevant characteristics of TiO2, IMAC, and antibody enrichment into a single structured comparison, helping researchers align method choice with study goals and sample complexity.
Table 1. TiO2 vs IMAC vs Antibody Phosphopeptide Enrichment Methods: Principles, Strengths, and Best-Use Scenarios
|
Comparison Dimension |
TiO2 (MOAC) |
IMAC |
Antibody-Based Immunoaffinity |
|
Core principle / binding mechanism |
Phosphate groups interact with TiO2 surface via Lewis acid–base interactions; binds in acidic buffer and elutes under basic conditions |
Phosphate groups bind chelated metal ions (e.g., Fe3+, Ti4+) via electrostatic and coordination interactions; typically eluted by pH shift or competitive reagents |
Phosphorylation-specific antibodies recognize epitopes and form immune complexes; capture via magnetic beads or agarose and release by elution |
|
Key component features |
High affinity for multi-phosphorylated peptides; exclusion-agent optimization reduces non-specific binding |
Metal ion choice is tunable; often enriches mono-phosphorylated peptides; metal ion selection can shift enrichment behavior |
Highly specific for defined phosphorylation types (e.g., pY); typically narrower coverage but very high purity |
|
Typical application range |
Large-scale phosphoproteomics of complex samples (e.g., cell or tissue lysates); compatible with automation platforms |
Protein complex- or interaction-adjacent phosphoproteomics; commonly used as a complementary method to expand coverage |
Signaling pathway studies (e.g., tyrosine kinase pathways); targeted clinical biomarker verification and validation workflows |
|
Key advantages |
High recovery potential; strong performance for multi-phosphorylated peptides; cost-effective and easy to scale/automate |
Flexible via metal-ion chemistry; good coverage for mono/basic phosphopeptides; compatible with downstream LC–MS quantification |
Highest specificity for the targeted phosphorylation class; strong interpretability for pathway-centric questions |
|
Key disadvantages |
Non-specific adsorption can occur without proper exclusion agents; often less coverage for basic phosphopeptides |
Recovery can be moderate; performance may be affected by co-enrichment of acidic peptides if conditions are not optimized |
Lower recovery and limited breadth; higher cost and potential batch-to-batch variability |
|
Core considerations / best-use scenarios |
Often the first choice for high-throughput discovery phosphoproteomics in complex matrices; well suited for standardized, scalable pipelines |
Best used alone when mono-phosphorylation/basic peptide capture is desired, or combined with TiO2 to broaden phosphosite coverage |
Best for targeted phosphoproteomics (e.g., pY-focused signaling) and clinical translation where specificity outweighs maximal site count |
Partner with MetwareBio for Comprehensive Phosphoproteomics Analysis
If your research depends on site-level phosphorylation quantification with strong depth and confident localization, MetwareBio offers a streamlined, end-to-end phosphoproteomics service designed for both discovery studies and translational applications. Our current workflow uses Ti-IMAC phosphopeptide enrichment combined with a 4D label-free strategy on the Bruker timsTOF HT platform, enabling sensitive and scalable phosphosite profiling across complex biological matrices.
What you gain by partnering with MetwareBio:
- High-efficiency Ti-IMAC phosphopeptide enrichment
A robust enrichment workflow optimized to increase phosphopeptide depth and reduce background interference, supporting reliable phosphosite identification in complex samples.
- 4D label-free phosphoproteomics on timsTOF HT
Ion mobility–enabled 4D data acquisition improves peptide separation and supports deeper, more consistent phosphoproteome coverage across large sample sets.
- Broad sample compatibility (human, animal, and plant)
We routinely process diverse matrices and tailor upstream preparation (extraction, digestion, enrichment conditions) to your sample type to protect phosphorylation information and maximize usable signal.
- Site-level quantification with biologically interpretable outputs
Deliverables emphasize phosphorylation at the site level, enabling differential phosphorylation analysis and pathway-level interpretation aligned with signaling biology.
- From sample to bioinformatics: a complete workflow
We support the full project lifecycle—sample processing, LC–MS/MS, quality control, statistical analysis, functional enrichment, and visualization—so you receive results that are ready for publication and follow-up validation.
- Experienced team and standardized execution
Our team brings extensive project experience and a standardized workflow to support reproducible data generation, with clear documentation and reporting.
To explore specifications, deliverables, and submission guidance, please refer to our phosphoproteomics services webpage.
References
1. Thingholm TE, Jensen ON, Larsen MR. Analytical strategies for phosphoproteomics. Proteomics. 2009;9(6):1451-1468. doi:10.1002/pmic.200800454
2. Leitner A. Phosphopeptide enrichment using metal oxide affinity chromatography. Trends Analyt Chem. 2016;82:457-467. doi:10.1016/j.trac.2016.06.012
3. Abelin JG, Trantham PD, Penny SA, et al. Complementary IMAC enrichment methods for HLA-associated phosphopeptide identification by mass spectrometry. Nat Protoc. 2015;10(9):1308-1318. doi:10.1038/nprot.2015.086
4. Yue X, et al. Comparing Multi-Step IMAC and Multi-Step TiO2 Methods for Phosphopeptide Enrichment. J Proteome Res. 2015;14(11):4833-4841. doi:10.1021/acs.jproteome.5b00617
5. Aryal UK, Ross AR. Enrichment and analysis of phosphopeptides under different experimental conditions using titanium dioxide affinity chromatography and mass spectrometry. Rapid Commun Mass Spectrom. 2010;24(2):219-231. doi:10.1002/rcm.4377
6. Kim J, et al. Comprehensive Evaluation of Different TiO2-Based Phosphopeptide Enrichment and Fractionation Methods for Phosphoproteomics. Cells. 2022;11(13):2047. doi:10.3390/cells11132047
7. Leitner A, et al. Robust, Sensitive, and Automated Phosphopeptide Enrichment Optimized for Low Sample Amounts Applied to Primary Hippocampal Neurons. J Proteome Res. 2016;15(11):3776-3787. doi:10.1021/acs.jproteome.6b00753
8. Gronauer TF, et al. Phosphopeptide Enrichment for Phosphoproteomic Analysis - A Tutorial and Review of Novel Materials. Anal Chem. 2020;92(1):26-34. doi:10.1021/acs.analchem.9b04629
9. Tape CJ, et al. Multiplexed Phosphoproteomic Profiling Using Titanium Dioxide and Immunoaffinity Enrichments Reveals Complementary Phosphorylation Events. J Proteome Res. 2017;16(4):1506-1514. doi:10.1021/acs.jproteome.6b00987
10. Korff K, et al. Zirconium(IV)-IMAC for phosphopeptide enrichment in phosphoproteomics. EMBO Mol Med. 2025;17(11):3174-3196. doi:10.1038/s44321-025-00309-0
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


