Unlocking the Lactylome: Bridging Metabolism, Epigenetics, and Gene Expression through Protein Lactylation
For decades, lactate was regarded as a metabolic byproduct of glycolysis. Advances in metabolomics, epigenetics, and mass spectrometry–based proteomics have redefined lactate as an active metabolic signal that regulates gene expression, immune responses, and cell fate decisions. The discovery of protein lactylation, particularly lysine lactylation (Kla), revealed a direct molecular link between cellular metabolism and epigenetic regulation. As a newly recognized post-translational modification, lactylation provides critical insights into cancer, inflammation, neurobiology, and metabolic diseases.
In this article, we present a concise overview of protein lactylation, including its biochemical mechanisms, biological functions, analytical approaches in lactylation proteomics, and emerging applications, highlighting its role in metabolic–epigenetic crosstalk.
What Is Protein Lactylation? A Metabolism-Driven Post-Translational Modification
Protein lactylation represents a paradigm shift in how cellular metabolism regulates protein function and gene expression. This chapter outlines the conceptual transition of lactate from a metabolic byproduct to a signaling molecule, introduces the discovery of lysine lactylation (Kla), and explains how lactylation functions as a direct metabolic–epigenetic feedback mechanism.
Lactate as a Signaling Metabolite
Historically, lactate was viewed as a byproduct of anaerobic glycolysis when pyruvate could not enter the tricarboxylic acid (TCA) cycle, associated with hypoxia, muscle fatigue, and pathological states such as cancer. However, metabolic studies later revealed that lactate is continuously produced and utilized even under normoxic conditions, indicating a more active biological role. Advances in systems biology and metabolomics have established lactate as a signaling metabolite involved in redox regulation, immune cell behavior, and intercellular communication. These findings laid the foundation for understanding how lactate can directly influence molecular and regulatory processes beyond classical metabolism.
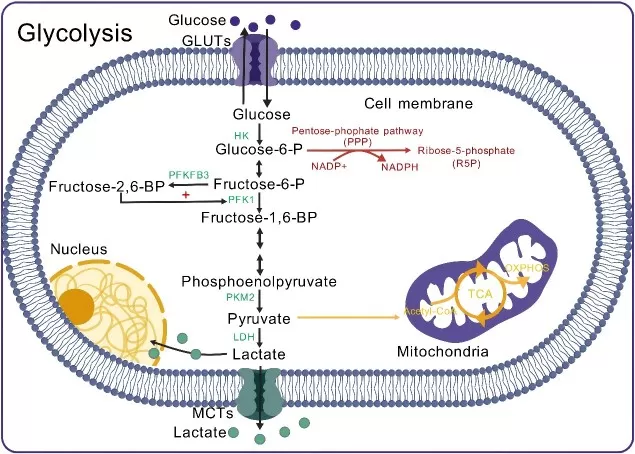
Aerobic glycolysis in immune cells
Image reproduced from Liu et al., 2025, Frontiers in immunology, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
Discovery of Lysine Lactylation (Kla)
In 2019, a landmark Nature study by Zhang et al. identified lysine lactylation (Kla) as a novel post-translational modification on histone proteins. Using mass spectrometry–based proteomics, the study demonstrated that lactate-derived lactyl groups can be covalently attached to lysine residues, directly affecting chromatin structure and gene transcription. This discovery established protein lactylation as a bona fide PTM and expanded the repertoire of metabolite-driven acyl modifications such as acetylation, methylation, and succinylation. Importantly, lysine lactylation (Kla) was shown to be dynamically regulated by intracellular lactate levels, providing a direct link between metabolic flux and epigenetic regulation.
Protein Lactylation as a Metabolic–Epigenetic Feedback Mechanism
At its core, protein lactylation functions as a metabolic feedback mechanism, enabling cells to sense and respond to changes in glycolytic activity. Under conditions such as the Warburg effect—where cancer cells preferentially rely on aerobic glycolysis—excess lactate accumulation drives the formation of lactyl-CoA, which subsequently serves as a substrate for lysine lactylation. Through this process, elevated glycolytic flux is converted into stable chemical marks on proteins, particularly histones, resulting in sustained changes in gene expression programs.
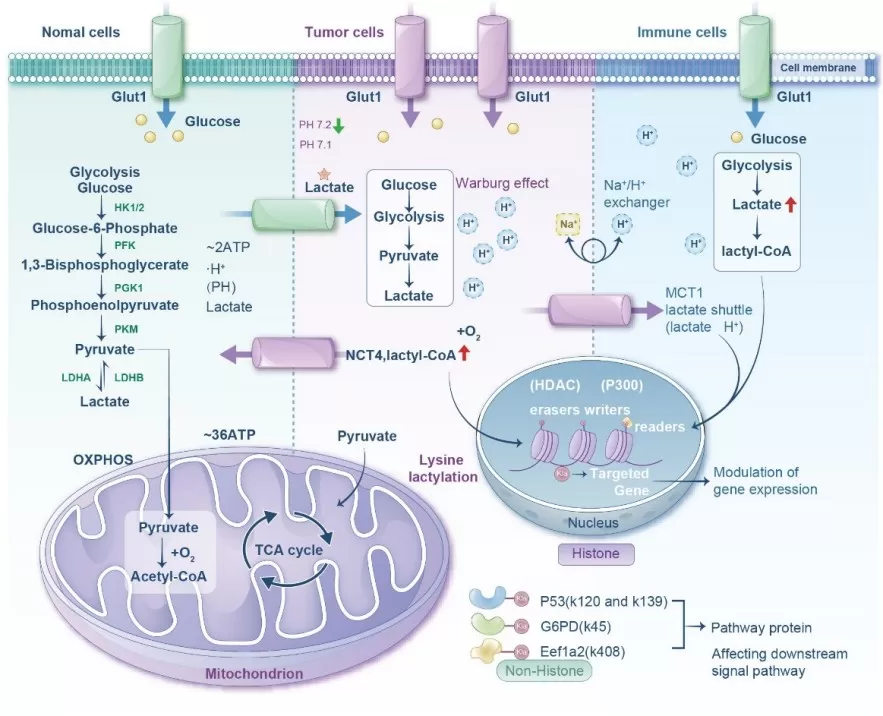
Diagram comparing metabolic processes in normal, tumor, and immune cells
Image reproduced fromYang et al., 2025, Frontiers in immunology, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
This mechanism provides a direct molecular explanation for how metabolic reprogramming can reshape transcriptional landscapes without requiring intermediary signaling cascades. Beyond histones, emerging evidence indicates that non-histone protein lactylation can modulate enzyme activity, protein stability, and subcellular localization, further extending the functional scope of lactylation. Together, these findings position protein lactylation as a central regulatory node at the intersection of metabolism, epigenetics, and cellular function.
The Biochemistry of Protein Lactylation: Donors, Writers, Erasers, and Substrates
Protein lactylation is a tightly regulated enzymatic process rather than a passive metabolic byproduct. Its establishment, removal, and functional consequences rely on specific biochemical components that translate intracellular lactate levels into regulatory signals. This chapter summarizes the core molecular machinery governing protein lactylation, including its metabolic donor, enzymatic writers and erasers, and major substrates.
Donors of Protein Lactylation: Lactyl-CoA as the Fuel for Lactylation
At the molecular level, lysine lactylation (Kla) involves the covalent attachment of a lactyl group to lysine residues, neutralizing their positive charge and altering protein function. Current evidence indicates that lactyl-CoA, derived from lactate metabolism, serves as the acyl donor for enzymatic lactylation. Although lactyl-CoA is less abundant than acetyl-CoA, its levels increase under enhanced glycolysis, hypoxia, inflammation, and the Warburg effect, explaining the strong metabolic sensitivity of protein lactylation and positioning it among metabolism-dependent post-translational modifications.
Writers of Protein Lactylation: p300 as a Lactyltransferase
Among the enzymes implicated in protein lactylation, p300 (EP300) is currently the best-characterized and most widely accepted lactyltransferase. Originally identified as a histone acetyltransferase, p300 exhibits remarkable catalytic flexibility and is capable of transferring multiple acyl groups to lysine residues, including acetyl, propionyl, crotonyl, and lactyl groups. This promiscuity allows p300 to function as a metabolic sensor that directly links intracellular acyl-CoA availability to chromatin modification.
Biochemical and structural studies indicate that p300 can utilize lactyl-CoA to catalyze lysine lactylation, particularly on histones at transcriptionally active regions. Elevated lactate production biases p300 activity toward lactylation, reshaping gene expression programs in highly glycolytic cells such as macrophages and tumor cells. While p300 is currently the primary known writer, the existence of additional lactyltransferases—especially for non-histone substrates—remains an important open question in lactylation biochemistry.
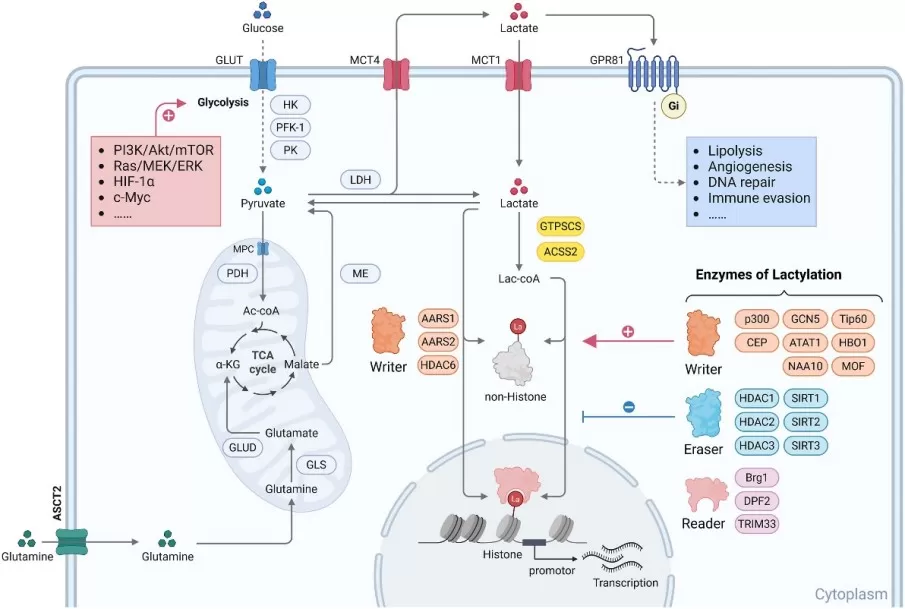
Overview of L-lactate metabolism and L-lactylation
Image reproduced from Zhang et al., 2025, Genes & Diseases, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
Erasers of Protein Lactylation: HDACs and Sirtuins in Delactylation
Protein lactylation is reversible and dynamically regulated by enzymes with delactylase activity. Several histone deacetylases, including HDAC1, HDAC2, and HDAC3, can remove lactyl groups from lysine residues, reflecting the mechanistic overlap among lysine acyl modifications. These enzymes play a crucial role in maintaining lactylation homeostasis and ensuring the temporal control of lactylation-dependent gene regulation. In addition, sirtuins (SIRT1, SIRT2, and SIRT3) contribute to delactylation in an NAD⁺-dependent manner, directly linking lactylation dynamics to cellular redox state, energy metabolism, and mitochondrial function within broader metabolic regulatory networks.
Substrates of Protein Lactylation: Histone vs. Non-Histone Kla
Histones were the first identified substrates of protein lactylation, establishing histone lactylation as a novel epigenetic mark involved in transcriptional regulation, particularly during inflammation resolution and tissue repair. Beyond histones, non-histone protein lactylation is now recognized as widespread, affecting metabolic enzymes, transcription factors, cytoskeletal proteins, and signaling molecules. These modifications regulate enzyme activity, protein stability, and protein–protein interactions in a context-dependent manner, greatly expanding the functional scope of the lactylome across cell types, metabolic states, and disease conditions.
Comparison of Histone Lactylation and non-Histone Lactylation (Reproduced from Peng and Du, 2025, Molecular biomedicine)
|
Aspect |
Histone lactylation |
non-histone lactylation |
|
Target Proteins |
Primarily occurs on histones H2A, H2B, H3, and H4 |
Found on various enzymes, transcription factors, and DNA damage repair proteins |
|
Number of Modification Sites |
Limited (28 sites) due to structural constraints |
Exceeds 9,000 lactylation sites according to current literature |
|
Type of Reaction |
Enzymatic reaction induced by L-lactate |
Enzymatic or non-enzymatic reactions induced by L-lactate or D-lactate |
|
Functional Mechanism |
Alters chromatin conformation to regulate gene transcription |
Modulates enzymatic activity, affects protein function, and reshapes protein interaction networks |
|
Biological Significance |
Primarily involved in epigenetic regulation |
Implicated in diverse cellular processes such as metabolism, DNA repair, and signal transduction |
Biological Functions of Protein Lactylation in Metabolic and Cellular Regulation
Protein lactylation has emerged as a key mechanism by which cells integrate metabolic information into functional outcomes. Rather than acting as a static modification, lactylation dynamically regulates protein behavior, metabolic responses, and cellular programs in accordance with intracellular lactate levels. In this chapter, we focus on the biological functions of protein lactylation at the molecular, metabolic, and cellular levels.
Protein Lactylation as a Modulator of Protein Function
Lactylation does not just "mark" a protein; it fundamentally alters its biochemical properties. By attaching a 3-carbon lactyl group to lysine residues, this modification can:
i. Alter Enzymatic Activity: Lactylation can stericallly hinder active sites or induce conformational changes, either activating or inhibiting metabolic enzymes.
ii. Regulate Protein Stability: Modification of specific lysine residues can prevent or facilitate degradation pathways (e.g., the ubiquitin-proteasome system).
iii. Change Subcellular Localization: Lactylation can act as a molecular "address," shifting proteins between the cytoplasm, nucleus, or mitochondria to meet cellular demands.
Protein Lactylation as a Metabolic Sensor and Effector
Protein lactylation is tightly coupled to cellular metabolic states, particularly enhanced glycolysis. Increased glycolytic flux leads to lactate accumulation and elevated lactyl-CoA availability, resulting in widespread lysine lactylation on both histone and non-histone proteins. This process enables lactylation to function as a metabolic sensor, translating energy availability into molecular regulation. Metabolic contexts associated with increased lactylation include aerobic glycolysis, hypoxia-driven metabolic adaptation, and high metabolic demand states, positioning lactylation as a direct effector linking metabolism to gene regulation and protein function.
Lactylation in Cellular State Transitions and Functional Programs
At the cellular level, protein lactylation acts as a regulatory mechanism that stabilizes transitions between functional states under sustained metabolic conditions. Persistent lactate accumulation induces coordinated changes across the lactylome, with histone lactylation reinforcing transcriptional programs linked to prolonged metabolic activity and non-histone lactylation modulating the activity and interactions of metabolic enzymes and signaling proteins. Through this integrated regulation, lactylation enables cells to align gene expression, enzymatic function, and signaling behavior with persistent energetic demands, supporting longer-term functional reprogramming without permanent genetic alteration.
Crosstalk Between Protein Lactylation and Other Post-Translational Modifications
Protein lactylation competes with other post-translational modifications (PTMs), particularly acetylation, for the same lysine residues. Both modifications neutralize the positive charge of lysine, but lactylation introduces a distinct metabolic signal derived from lactate metabolism, while acetylation is more closely associated with transcriptional activation. Enzymes such as p300 and HDACs play a key role in catalyzing both lactylation and acetylation, making their competition especially relevant under conditions of altered glycolytic flux, where lactyl-CoA availability may influence whether lactylation or acetylation predominates. This competition helps cells fine-tune chromatin states and gene expression patterns based on metabolic inputs.
Beyond competition, protein lactylation can cooperate with other PTMs to regulate protein function and cellular processes. Crosstalk among PTMs enables proteins to integrate signals from metabolism, stress, and nutrient availability, leading to precise cellular responses. Key features of PTM crosstalk involving lactylation include:
a) Sequential modifications: Multiple PTMs may occur on the same protein, influencing its function.
b) Synergistic regulation: Lactylation can work with other modifications like acetylation and methylation to regulate transcription factors and metabolic enzymes.
c) Context-dependent switching: The combination of PTMs changes based on metabolic shifts, allowing cells to adapt to environmental and internal signals.
These interactions allow lactylation to be a key player in the dynamic regulation of cellular processes, responding to diverse metabolic and environmental cues.
Technical Workflow: How to Study Lactylation and the Lactylome
Studying protein lactylation requires specialized experimental and analytical strategies that account for its metabolic sensitivity and relatively low abundance. Modern lactylomics relies on a combination of antibody-based assays, PTM-optimized mass spectrometry, and metabolic labeling approaches to achieve accurate and comprehensive characterization of the lactylome.
Antibody-Based Approaches: Pan-Kla Detection and Chromatin Analysis
Pan-lysine lactylation (Pan-Kla) antibodies provide a practical entry point for studying protein lactylation at both the global and locus-specific levels. In standard workflows, Pan-Kla antibodies are used in Western blotting to assess overall lactylation levels across different conditions, such as metabolic stress or immune activation. When combined with chromatin immunoprecipitation followed by sequencing (ChIP-seq), these antibodies enable genome-wide mapping of histone lactylation, revealing how lactylation redistributes across regulatory regions.
Although antibody-based methods lack site-level resolution, they remain essential for hypothesis generation, validation experiments, and linking lactylation changes to transcriptional outcomes in lactylomics studies.
Lactylation Proteomics: PTM-Optimized Mass Spectrometry for the Lactylome
Lactylation proteomics based on high-resolution mass spectrometry is the gold standard for global lactylation profiling. In these workflows, lactylated peptides are selectively enriched using antibody-based affinity purification, followed by LC–MS/MS analysis. This approach enables site-specific identification and quantification of lactylation across the entire lactylome, including both histone and non-histone proteins.
Advanced platforms such as timsTOF HT offer distinct advantages for lactylomics, including high sensitivity, fast acquisition speed, and improved separation of isobaric and near-isobaric PTMs. These capabilities are particularly important for distinguishing lysine lactylation from chemically similar acyl modifications, ensuring confident annotation of lactylation sites in complex biological samples.
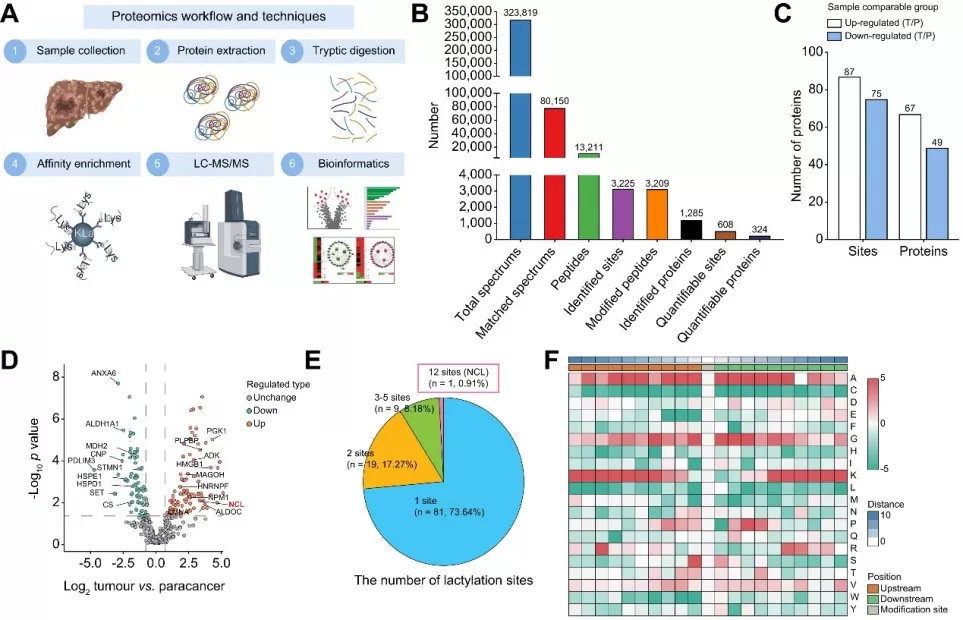
iCCA cells display a widely aberrant landscape of protein lactylation via MS-based lactylation profiling.
Image reproduced from Yang et al., 2024, Journal of hepatology, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
Metabolic Labeling: Tracing the Origin of Lactylation
Metabolic labeling strategies provide direct evidence linking cellular metabolism to protein lactylation. By culturing cells with ¹³C-labeled glucose or lactate, researchers can trace the incorporation of labeled carbon atoms into lactyl-CoA and subsequently into lactylated peptides. When combined with lactylation proteomics, this approach enables precise determination of lactyl group origin and turnover dynamics.
Metabolic labeling is particularly valuable in lactylomics studies focused on metabolic–epigenetic crosstalk, as it distinguishes lactylation driven by endogenous glycolysis from extracellular lactate uptake. This strategy strengthens causal interpretations and provides mechanistic insight into how metabolic flux shapes the lactylome under physiological and pathological conditions.
Cutting-Edge Applications of Protein Lactylation in Disease and Metabolism
As research on protein lactylation has rapidly expanded, its biological significance is increasingly being validated across diverse disease contexts. Advances in lactylation proteomics and global lactylation profiling have enabled researchers to move beyond mechanistic discovery toward functional and translational applications. In this section, we highlight representative case studies that illustrate how lactylation integrates metabolic reprogramming with disease progression in cancer, neurological systems, and inflammatory disorders.
Protein Lactylation in Cancer Metabolism and Tumor Progression
Aberrant glucose metabolism and lactate accumulation are hallmarks of cancer, and recent studies indicate that protein lactylation serves as a key molecular mechanism translating these metabolic changes into oncogenic regulation. Beyond histone modification, emerging evidence shows that non-histone lactylation can directly modulate RNA processing, signaling pathways, and tumor cell behavior, expanding the functional landscape of the cancer lactylome.
A recent study on intrahepatic cholangiocarcinoma (iCCA) demonstrated that glycolysis-derived lactate induces p300-mediated lactylation of nucleolin (NCL). Using lactylation-focused proteomics, site-specific validation, and functional assays, the authors showed that NCL lactylation alters alternative splicing of MADD, leading to sustained ERK signaling and enhanced tumor growth. This work reveals a previously unrecognized lactylation–RNA splicing axis in cancer metabolism, highlighting lactylation as both a mechanistic driver of tumor progression and a potential therapeutic target.
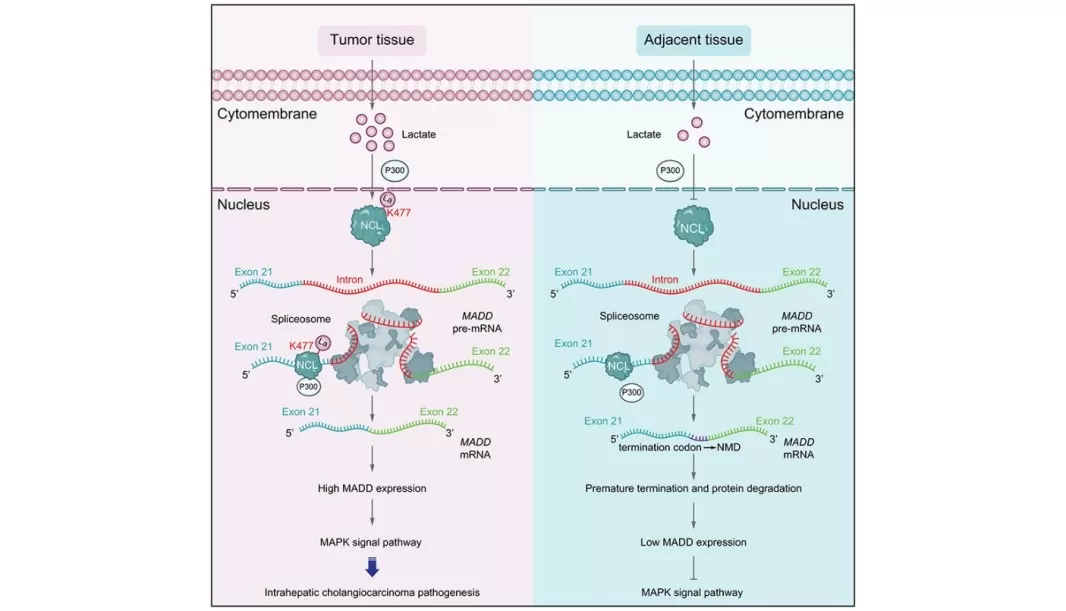
Glycolysis-Driven Nucleolin Lactylation Promotes Intrahepatic Cholangiocarcinoma Progression via MADD-Dependent ERK Activation
Image reproduced from Yang et al., 2024, Journal of hepatology, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
Lactylation in Brain Metabolism and Neural Function
Emerging evidence indicates that lactate is an active metabolic signal in the brain, coupling neuronal energy metabolism to epigenetic regulation. In the adult hippocampus, dynamic metabolic communication between neural cells supports neurogenesis, and histone lactylation has recently been recognized as a key molecular link connecting lactate flux to gene expression programs that govern neural stem cell fate.
In a mouse study titled Lactate shuttling links histone lactylation to adult hippocampal neurogenesis, researchers demonstrated that lactate transport and intracellular availability directly regulate histone lactylation levels in neural stem and progenitor cells. Using genetic and pharmacological modulation of lactate shuttling, combined with histone lactylation analysis and transcriptional profiling, the study showed that increased histone lactylation activates gene programs promoting hippocampal neurogenesis. These findings establish lactylation as a metabolic–epigenetic mechanism in brain plasticity, highlighting its importance in neurodevelopment and neurological health.
Lactylation in Inflammation and Immune Regulation
Inflammation and immune responses are tightly coupled to metabolic reprogramming, particularly enhanced glycolysis and lactate accumulation. Growing evidence indicates that protein lactylation, especially histone lactylation, functions as a central regulatory mechanism that converts inflammatory metabolic stress into immune and transcriptional regulation. Through this pathway, lactylation shapes gene expression programs that influence inflammatory intensity, immune cell behavior, and disease progression.
In a recent research article titled Lactate and H3K18 Lactylation Contribute To the Exacerbation of Acute Pancreatitis by Modulating NCOA4-mediated Ferroptosis, the authors demonstrated that elevated lactate levels in acute pancreatitis drive H3K18 lactylation in pancreatic cells. Using histone lactylation analysis, gene expression profiling, and functional ferroptosis assays, they showed that H3K18 lactylation upregulates NCOA4-mediated ferritinophagy, promoting ferroptotic cell death and exacerbating pancreatic injury. This study provides direct experimental evidence that lactylation links inflammatory metabolism to cell death pathways, highlighting histone lactylation as a potential therapeutic target in inflammation-associated diseases.
Challenges and Future Frontiers of Protein Lactylation Research
Despite rapid advances in protein lactylation research, several fundamental challenges remain unresolved. One of the most critical questions concerns site specificity—why certain lysine residues are preferentially lactylated while others remain unmodified under similar metabolic conditions. This selectivity likely reflects a combination of local protein structure, chromatin accessibility, and differential affinity of enzymes such as p300 for specific substrates. Addressing this challenge will require systematic lactylome-wide profiling combined with structural biology and quantitative lactylomics approaches.
Another major frontier lies in the identification of additional cofactors and regulatory enzymes involved in lactylation. While p300, HDACs, and sirtuins are currently the best-characterized writers and erasers, the existence of lactylation-specific readers remains largely unexplored. Discovering these readers is essential for understanding how lactylation signals are interpreted at the molecular level and integrated into broader post-translational modification networks. Advances in proteomics, interactomics, and functional screening are expected to accelerate progress in this area.
Finally, the clinical translation of protein lactylation represents a promising yet challenging direction. Targeting lactylation-related pathways—such as lactate metabolism or p300-mediated lactylation—offers new opportunities for therapeutic intervention in cancer and inflammatory diseases. However, translating these strategies into clinical practice will require improved specificity, robust biomarkers, and a deeper understanding of lactylation dynamics in human tissues. Together, these efforts will define the next phase of lactylation research and its impact on precision medicine.
Reference
1. Liu, W., Yang, R., Zhan, Y., Yang, X., Zeng, H., Chen, B., Zeng, J., Hu, T., Hu, J., Xiao, Q., Shao, Y., & Chen, X. (2025). Lactate and lactylation: emerging roles in autoimmune diseases and metabolic reprogramming. Frontiers in immunology, 16, 1589853. https://doi.org/10.3389/fimmu.2025.1589853
2. Zhang, D., Tang, Z., Huang, H., Zhou, G., Cui, C., Weng, Y., Liu, W., Kim, S., Lee, S., Perez-Neut, M., Ding, J., Czyz, D., Hu, R., Ye, Z., He, M., Zheng, Y. G., Shuman, H. A., Dai, L., Ren, B., Roeder, R. G., … Zhao, Y. (2019). Metabolic regulation of gene expression by histone lactylation. Nature, 574(7779), 575–580. https://doi.org/10.1038/s41586-019-1678-1
3. Yang B, Li L, Shi D, Zhong T and Xiong H (2025) Lactylation and antitumor immunity. Frontiers in immunology, 16:1690068. https://doi.org/10.3389/fimmu.2025.1690068
4. Zhang, W., Huang, G., Tang, W., Li, J., Chen, J., Feng, Y., Li, K., Pan, C., Li, S., Zhang, H., Ye, R., Long, H., & Yi, G.-z. (2025). Lactylation-driven therapeutic resistance in cancer: Mechanisms and therapeutic opportunities. Genes & Diseases, 101935. https://doi.org/10.1016/j.gendis.2025.101935
5. Peng, X., & Du, J. (2025). Histone and non-histone lactylation: molecular mechanisms, biological functions, diseases, and therapeutic targets. Molecular biomedicine, 6(1), 38. https://doi.org/10.1186/s43556-025-00275-6
6. Yang, L., Niu, K., Wang, J., Shen, W., Jiang, R., Liu, L., Song, W., Wang, X., Zhang, X., Zhang, R., Wei, D., Fan, M., Jia, L., & Tao, K. (2024). Nucleolin lactylation contributes to intrahepatic cholangiocarcinoma pathogenesis via RNA splicing regulation of MADD. Journal of hepatology, 81(4), 651–666. https://doi.org/10.1016/j.jhep.2024.04.010
7. Li, Z., Liang, Z., Qi, H., Luo, X., Wang, M., Du, Z., & Guo, W. (2025). Lactate shuttling links histone lactylation to adult hippocampal neurogenesis in mice. Developmental cell, 60(8), 1271–1273. https://doi.org/10.1016/j.devcel.2025.01.007
8. Han, Z. Y., Ma, X. Y., Ma, S. Y., Shen, Z. J., Lu, Z. H., Sun, Y., & Yu, W. L. (2025). Lactate and H3K18 Lactylation Contribute To the Exacerbation of Acute Pancreatitis by Modulating NCOA4-mediated Ferroptosis. Inflammation, 10.1007/s10753-025-02361-x. Advance online publication. https://doi.org/10.1007/s10753-025-02361-x
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


