Proteomics in Drug Discovery: Accelerating Target Identification and Efficacy Evaluation
Why Proteomics Matters for Drug Discovery
Drug discovery has long leaned on genomic blueprints, yet proteins are the direct executors of cellular function—and they represent the primary binding partners or downstream effectors for most therapeutics. As noted in the literature, "proteins are the main targets of most drugs" [1]. However, genomics-first workflows often struggle to fully explain a drug’s mechanism of action (MoA), leaving gaps such as ambiguous pathways, hidden off-target interactions, and a shortage of pharmacodynamic (PD) or efficacy biomarkers that translate reliably into the clinic. Practical bottlenecks include suboptimal efficacy, off-target delivery, and the analytical burden of large, heterogeneous datasets spanning genomics and proteomics—factors that slow target identification and validation [2]. Proteomics helps close these gaps by offering a system-level view of disease phenotypes and how bioactive molecules reshape protein abundance, activity states, and network behavior, enabling higher-resolution interpretation of why and how drug responses occur [1]. Its impact has grown as the field has moved from small, single-cohort studies to large biobank-scale efforts, exemplified by the UK Biobank (>54,000 participants). Such initiatives profile plasma proteomes to reveal disease signatures and build predictive models, creating a rich foundation for biomarker discovery and therapeutic development [3]. By shifting attention from the "gene blueprint" to the "protein execution" layer, proteomics supports a transition from hypothesis-driven guesswork to evidence-based decision-making—reducing uncertainty while accelerating the identification of actionable, druggable targets.
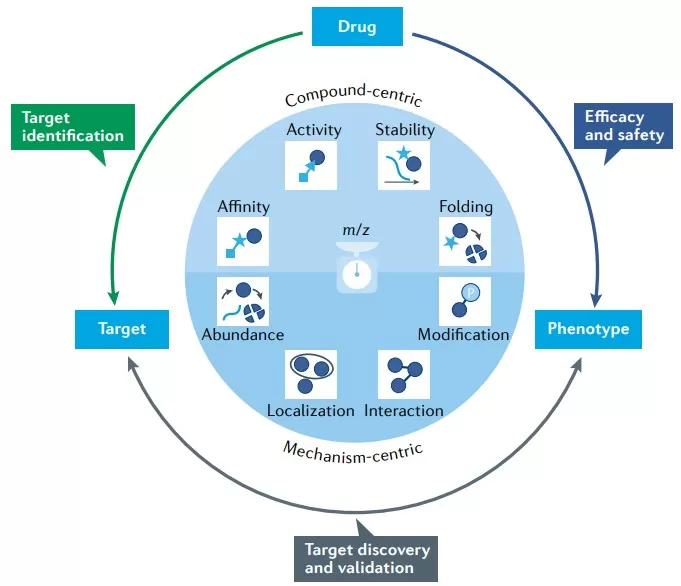
proteomics applications in the preclinical drugdiscovery process
Where Proteomics Fits in the Drug Discovery Pipeline
Proteomics contributes value across the entire drug discovery pipeline, providing decision-support data at each stage and helping teams move from observations to actionable mechanisms. In target identification and validation, approaches such as chemoproteomics enable unbiased mapping of protein targets while flagging potential safety hazards early. For example, pan-cancer analyses of druggable proteins across multiple tumor types can reveal dependencies driven by protein overexpression or hyperactivation, improving the accuracy of target prioritization and predicting which targets are most likely to yield therapeutic benefit [1, 4]. At this stage, proteogenomic integration further strengthens validation—for instance, combining proteomics with genomics to uncover synthetic-lethality strategies linked to tumor suppressor loss [4]. During hit-to-lead optimization, proteomics informs molecule selection by assessing protein affinity, functional activity, and stability, thereby clarifying whether a candidate modulates the intended biology. Emerging strategies such as targeted protein degradation and reactive-fragment screening add experimental leverage and guide study design so that lead compounds more consistently drive measurable phenotypic modulation [1]. In lead optimization and preclinical development, proteomics supports candidate refinement by surfacing off-target effects and safety liabilities before they become costly failures. In clinical phases, proteomic readouts can be used for PD monitoring, patient stratification, and resistance mechanism analysis. Tumor proteomics, for example, can augment conventional risk scores and enable companion-diagnostic development for personalized therapy, as illustrated by resources such as the LinkedOmics database [3-7]. Taken together, this end-to-end integration turns proteomic measurements into a continuous evidence stream that helps convert raw data into actionable drug candidates.
Method Toolbox: From Detecting Proteins to Demonstrating Drug Action
Proteomics provides a versatile set of strategies to systematically evaluate how drugs affect proteins—capturing changes in affinity, activity, stability, folding, and broader network context—to support mechanism-of-action studies and target validation. Methods such as chemoproteomics can dissect disease phenotypes and quantify how they respond to bioactive molecules, helping to identify safety hazards as well as uncover novel therapeutic opportunities. For example, mass spectrometry-based workflows can track dynamic shifts in protein states after treatment, supporting prediction of efficacy and early detection of off-target effects. A key advantage is unbiased, system-wide profiling, which can reveal complex and previously hidden modulations at high resolution and improve target deconvolution. At the same time, these approaches can be technically demanding, and interpretation is susceptible to noise; robust experimental controls and advanced analytical pipelines are therefore required to separate true biological signals from artifacts. [1]
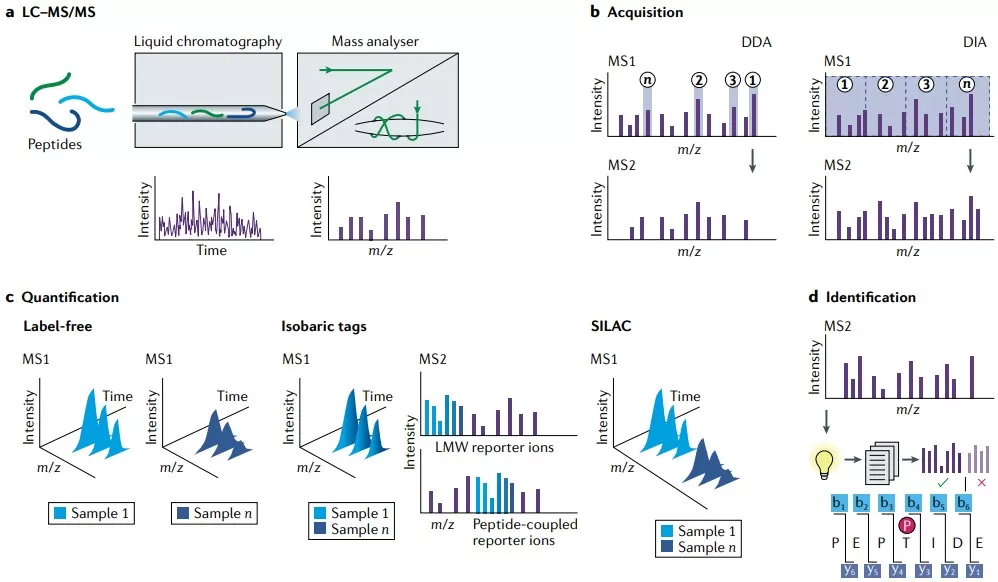
Major protein identification and quantification strategies
1) Quantitative proteomics (DIA, TMT, SOMAscan, Olink)
Techniques such as Data-Independent Acquisition (DIA), Tandem Mass Tag (TMT), SOMAscan, and Olink enable broad, quantitative measurement of the proteome at scale, making them central to large-cohort profiling in drug discovery. They support target identification by revealing disease-associated protein abundance patterns—such as pan-cancer analyses that highlight overexpressed proteins linked to druggable dependencies. Key advantages include high throughput and extensive coverage, as well as the ability to integrate proteomic measurements with genomic information for more accurate target prioritization; for example, studies that combine proteomic data with genetic screens can better predict effective targets for therapy development. Important limitations include difficulty quantifying low-abundance proteins and susceptibility to sample-to-sample variability, both of which can distort mRNA–protein correlations and reduce reliability in heterogeneous settings such as tumors. [3-4]
2) PTM proteomics (phosphorylation, acetylation, etc.)
Post-translational modification (PTM) proteomics—focused on modifications such as phosphorylation and acetylation—captures functional protein regulation induced by drugs and helps explain mechanisms like kinase signaling rewiring or metabolic cross-talk. It can expose new drug opportunities: phosphoproteomics may uncover links between tumor suppressor loss and targetable kinases, while acetylproteome studies can highlight roles in DNA damage responses and cellular metabolism. Advantages include high specificity for dynamic, regulatory events that directly drive disease phenotypes, enabling more precise annotation of targetable pathways for personalized therapy. Limitations arise from the technical challenge of detecting transient or low-stoichiometry modifications and the need for specialized instrumentation and workflows, which can complicate scaling and undermine reproducibility across studies. [7]
3) Spatial proteomics for drug mechanism-of-action research
Spatial proteomics links protein localization and molecular interactions to cellular context, strengthening mechanism-of-action studies by connecting where proteins are in tissues to what they do under drug treatment. When combined with advanced imaging modalities and AI-enabled analysis, spatial proteomics can deliver relatively unbiased molecular readouts that sit close to biological function, offering insight into tissue-level responses. Its major advantage is the ability to resolve spatial heterogeneity—for example, within tumors—thereby clarifying how microenvironments influence drug distribution, target engagement, and efficacy, which is critical for precision medicine. Limitations include reliance on specialized platforms (e.g., multiplexed imaging, laser microdissection) that are resource-intensive and not always available for routine workflows, while the resulting high-dimensional data typically require sophisticated computational methods for interpretable conclusions. [6]
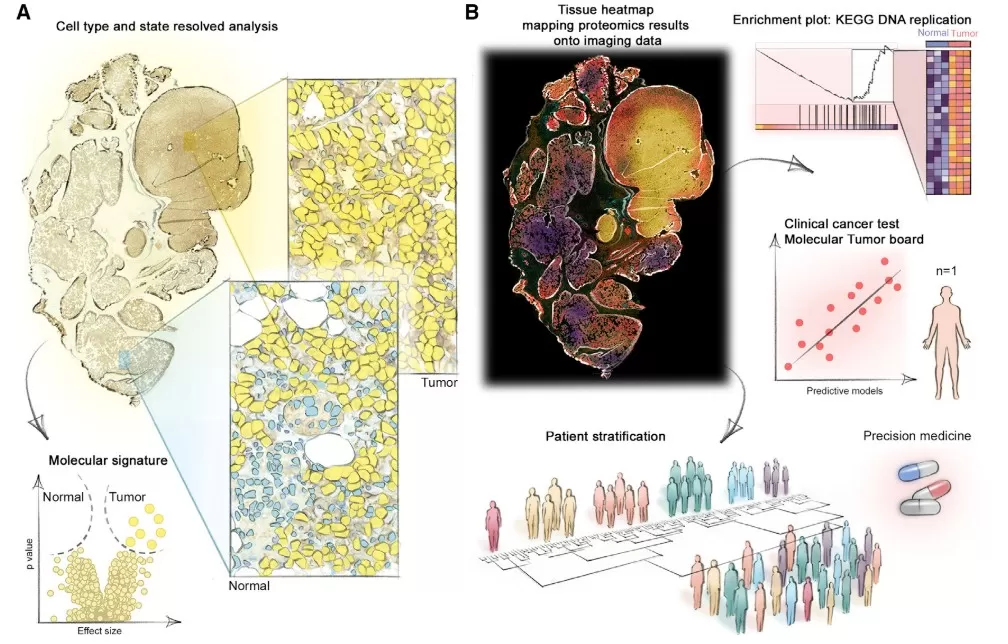
Clinical applications of spatial proteomics for patient phenotyping
4) Chemoproteomics and affinity enrichment (kinobeads, ABPP, etc.)
Chemoproteomics approaches—such as kinobeads and activity-based protein profiling (ABPP)—use affinity enrichment to map drug–protein interactions, supporting target deconvolution and validation of bioactive molecules. They are particularly useful in early discovery, where understanding target engagement and safety profiles can shape go/no-go decisions; for instance, reactive fragments can be used to probe protein activities and reveal tractable binding opportunities. Key advantages include sensitivity to low-affinity binders and applicability across diverse drug modalities, enabling rapid screening and identification of novel targets. Limitations include the risk of false positives due to non-specific binding and the need for careful probe design and optimization, which can restrict broad applicability and increase experimental costs. [1]
5) Intracellular target binding and occupancy: CETSA/TPP, LiP-MS, etc.
Methods such as the Cellular Thermal Shift Assay (CETSA), Thermal Proteome Profiling (TPP), and Limited Proteolysis–Mass Spectrometry (LiP-MS) assess intracellular target binding and occupancy by quantifying drug-induced changes in protein stability and conformation. Because they operate in live cells or native-like cellular environments, these approaches can provide direct evidence of target engagement that complements conventional biochemical assays and supports both safety hazard identification and mechanism-informed optimization. Their strengths include physiological relevance and the ability to capture binding events in complex cellular settings. Limitations include reduced sensitivity for weak or transient interactions and potential interference from cellular complexity, which can obscure specific binding signatures and often necessitates extensive follow-up validation.
6) Artificial intelligence and deep learning: proteomics for drug discovery
AI and deep learning methods can integrate proteomic datasets with other omics layers to accelerate drug discovery tasks such as virtual screening, toxicity prediction, and biomarker identification. By modeling relationships across genomic, proteomic, and transcriptomic features, these approaches can improve prediction of drug responses and sharpen target prioritization. Advantages include efficient handling of high-dimensional data and the potential to reduce time and cost through automated, scalable workflows—for example, in pharmacogenomics contexts where AI supports the development of targeted therapeutics. Limitations include dependence on large, high-quality training datasets (with the risk of bias or limited generalizability) and the “black-box” nature of many models, which can hinder interpretability and slow clinical translation. [2]
Summary of Proteomics Analysis Tools for Drug Discovery
|
Method/Tool |
Role in Drug Discovery |
Advantages |
Limitations |
|
Quantitative proteomics (DIA, TMT, SOMAscan, Olink) |
Large-scale protein quantification for target nomination and dependency analysis. |
High throughput; broad coverage; strong integration with genomics. |
Limited sensitivity for low-abundance proteins; batch effects. |
|
PTM proteomics (phosphorylation, acetylation) |
Detects drug-perturbed PTM signaling and actionable pathways. |
Captures dynamic regulation; enables pathway-level insights. |
Low-stoichiometry PTMs are challenging; complex workflows. |
|
Spatial proteomics |
Defines protein localization and interactions in tissue context. |
Resolves spatial heterogeneity and microenvironment effects. |
High cost; complex data analysis requirements. |
|
Chemoproteomics / affinity enrichment (kinobeads, ABPP) |
Identifies drug–protein interactions for target deconvolution. |
Sensitive to weak binders; supports early safety profiling. |
Non-specific binding; costly probe design. |
|
Intracellular target binding (CETSA, TPP, LiP-MS) |
Validates in-cell target engagement and occupancy. |
Physiologically relevant evidence of mechanism. |
May miss transient interactions; signal complexity. |
|
AI and deep learning |
Predictive modeling for targets, mechanisms, and biomarkers. |
Handles high-dimensional data efficiently. |
Data-hungry; limited interpretability. |
Accelerating Target Identification: From Phenotype to Mechanism (From Phenotype to Target/MoA)
Proteomics enables “unbiased” discovery that links phenotypes to mechanism, helping researchers pinpoint direct targets, map bypass pathways, detect off-target effects, and surface toxicity clues—thereby moving drug discovery from descriptive phenotypic observations to mechanistic understanding. When combined with proteogenomic integration, these datasets can support strategies such as synthetic lethality (to exploit tumor suppressor loss) and the prioritization of mutant peptides as neoantigens for immunotherapy [4]. Proteomics can also identify hyperactivation-driven dependencies, where protein overexpression correlates with drug sensitivity, enabling more precise predictions of tractable targets. Case Study 1: CPTAC Pan-Cancer Analysis - The Clinical Proteomic Tumor Analysis Consortium (CPTAC) integrated proteogenomic data from 1,043 patients spanning 10 cancer types and identified 2,863 druggable proteins, revealing a wide abundance range and biological factors that influence mRNA–protein correlations. By combining tumor proteomic measurements with genetic-screen data from cancer cell lines, the analysis highlighted protein overexpression-driven druggable dependencies (including kinase-associated vulnerabilities), accurately predicted effective targets, and nominated peptides for immunotherapy. Overall, this example illustrates how proteomics can expose mechanisms underlying both drug efficacy and resistance. [4]
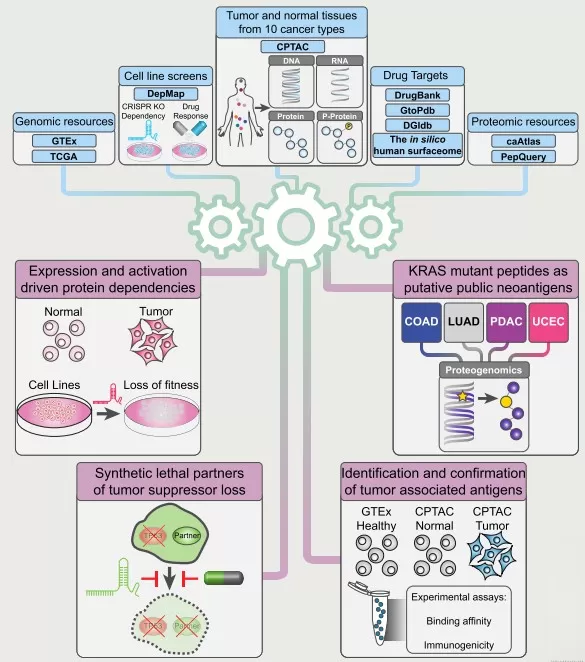
Pan-cancer proteogenomics expands the landscape of therapeutic targets
Efficacy Evaluation: From “Tumor Shrinkage” to “Pathway Switch”
Proteomics elevates efficacy assessment from coarse endpoints (e.g., tumor shrinkage) to molecular evidence that a pathway has been engaged and modulated, increasing confidence that a drug is acting on its intended biology. Target engagement can be supported by approaches such as CETSA, which infers binding by monitoring drug-induced thermal stability shifts in proteins [1]. Beyond binding, phosphoproteomics can quantify pathway modulation by profiling how signaling networks are rewired under treatment; for example, in breast cancer, phosphoproteomics revealed kinase associations that predict responsiveness to inhibitors such as CDK4/6, providing a dynamic readout of pathway activity [7]. Proteomic biomarkers can also enable predictive and companion diagnostics by identifying responder populations and clarifying resistance mechanisms. Proteogenomic analyses strengthen these links by connecting genetic variation to protein abundance and function, as in Mendelian-randomization studies implicating targets such as PCSK9 in cholesterol management or cytokines in inflammation [3, 8]. Case Study 2 (Proteogenomics in Liver Cancer) - In the Liver Cancer Organoid Biobank (LICOB) study, proteogenomic profiling of patient-derived organoids defined proliferative and metabolic subtypes associated with prognosis. High-throughput drug screening then revealed distinct response patterns tied to multi-omics signatures, enabling prediction of effective drug combinations. For example, synergistic inhibition of mTOR together with tyrosine kinase inhibitors was validated in model systems, demonstrating how proteomics can connect response phenotypes to the underlying molecular pathways and support personalized treatment strategies. The study also provides access via a dedicated web portal. [9]
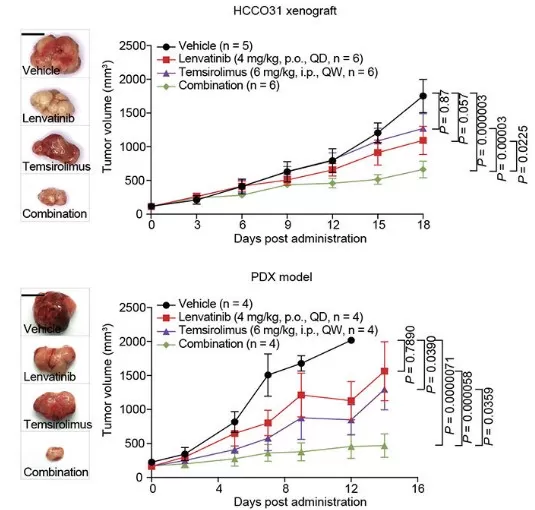
Drug Combination Prediction and Validation in LICOB
Implementation Advice: Designing Actionable Proteomics Studies
To make proteomics studies truly “decision-ready,” it helps to follow a structured workflow that begins with problem definition. First, articulate the drug discovery question in operational terms—for example, distinguishing on-target from off-target effects, identifying resistance biomarkers, or uncovering compensatory pathways. Next, design experiments that capture real-world variability, including sufficiently large cohorts (e.g., biobanks) when the goal is population-level inference. The Pharma Proteomics Project, for instance, applied standardized plasma proteomics protocols across 54,219 participants to support robust biological validation [3]. For analysis, integrate proteomics with genomics and transcriptomics using approaches such as pathway mapping and pQTL (protein quantitative trait loci) analysis to connect genetic variants with protein levels, thereby nominating biomarkers and drug targets [8, 10]. Orthogonal validation should then confirm key findings using independent methods—such as mouse knockdown experiments for trans-pQTL hypotheses or clinical-trial evidence for candidate chemokine receptors—so that associations translate into causal, testable claims [3, 8] Finally, translate discoveries into scalable assays (e.g., PRM/SRM or immunoassays) that can be deployed in preclinical and clinical settings. This final step is critical for operationalization, as exemplified by workflows in which proteogenomic insights are converted into companion diagnostics or targeted-therapy strategies [1, 4, 11]. By progressing from problem definition to deployable assay, proteomics studies are more likely to avoid common pitfalls and deliver actionable outputs for drug development.
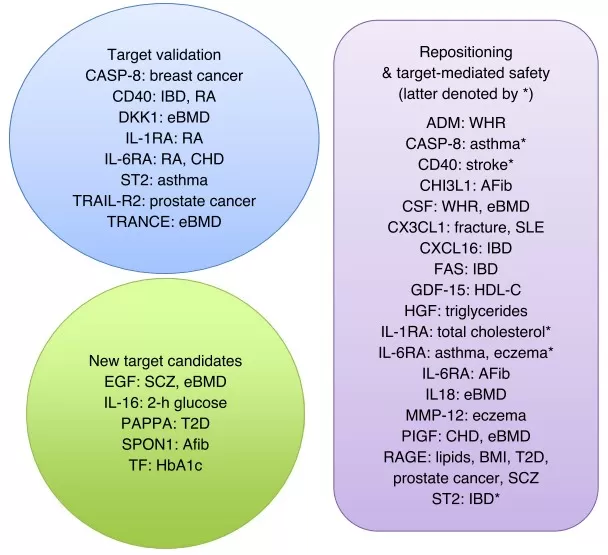
Protein–trait relationships that support target validation
Conclusion: Building Evidence Chains Instead of Guesswork
Proteomics is reshaping drug discovery by replacing conjecture with coherent evidence chains that connect targets, mechanisms, and outcomes. It clarifies drug mechanisms at the protein level, supports precise target validation, and enables efficacy prediction through biomarker-driven readouts—trends reinforced by ongoing advances in proteogenomics and AI-enabled integration. This shift not only accelerates target identification and efficacy evaluation, but also shows "much promise for future research" in areas such as stroke and myocardial infarction [2, 5]. By bridging genomic signals with protein-level biology and clinical outcomes, proteomics helps researchers develop therapies that are safer, more effective, and more translatable—moving the field from an art informed by intuition to a science guided by data.
References:
1. Meissner F, Geddes-McAlister J, Mann M, Bantscheff M. The emerging role of mass spectrometry-based proteomics in drug discovery. Nat Rev Drug Discov. 2022 Sep;21(9):637-654. doi: 10.1038/s41573-022-00409-3.
2. Gupta R, Srivastava D, Sahu M, Tiwari S, Ambasta RK, Kumar P. Artificial intelligence to deep learning: machine intelligence approach for drug discovery. Mol Divers. 2021 Aug;25(3):1315-1360. doi: 10.1007/s11030-021-10217-3.
3. Sun BB, Chiou J, Traylor M, Benner C, Hsu YH, Richardson TG, Surendran P, Mahajan A, Robins C, Vasquez-Grinnell SG, et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature. 2023 Oct;622(7982):329-338. doi: 10.1038/s41586-023-06592-6.
4. Savage SR, Yi X, Lei JT, Wen B, Zhao H, Liao Y, Jaehnig EJ, Somes LK, Shafer PW, Lee TD, Fu Z, Dou Y, Shi Z, Gao D, Hoyos V, Gao Q, Zhang B. Pan-cancer proteogenomics expands the landscape of therapeutic targets. Cell. 2024 Aug 8;187(16):4389-4407.e15. doi: 10.1016/j.cell.2024.05.039.
5. Webb RJ, Al-Asmakh M, Banach M, Mazidi M. Application of proteomics for novel drug discovery and risk prediction optimisation in stroke and myocardial infarction: a review of in-human studies. Drug Discov Today. 2024 Nov;29(11):104186. doi: 10.1016/j.drudis.2024.104186.
6. Mund A, Brunner AD, Mann M. Unbiased spatial proteomics with single-cell resolution in tissues. Mol Cell. 2022 Jun 16;82(12):2335-2349. doi: 10.1016/j.molcel.2022.05.022.
7. Krug K, Jaehnig EJ, Satpathy S, Blumenberg L, Karpova A, Anurag M, Miles G, Mertins P, Geffen Y, Tang LC, et al. Proteogenomic Landscape of Breast Cancer Tumorigenesis and Targeted Therapy. Cell. 2020 Nov 25;183(5):1436-1456.e31. doi: 10.1016/j.cell.2020.10.036.
8. Folkersen L, Gustafsson S, Wang Q, Hansen DH, Hedman ÅK, Schork A, Page K, Zhernakova DV, Wu Y, Peters J, et al. Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat Metab. 2020 Oct;2(10):1135-1148. doi: 10.1038/s42255-020-00287-2.
9. Ji S, Feng L, Fu Z, Wu G, Wu Y, Lin Y, Lu D, Song Y, Cui P, Yang Z, Sang C, Song G, Cai S, Li Y, Lin H, Zhang S, Wang X, Qiu S, Zhang X, Hua G, Li J, Zhou J, Dai Z, Wang X, Ding L, Wang P, Gao D, Zhang B, Rodriguez H, Fan J, Clevers H, Zhou H, Sun Y, Gao Q. Pharmaco-proteogenomic characterization of liver cancer organoids for precision oncology. Sci Transl Med. 2023 Jul 26;15(706):eadg3358. doi: 10.1126/scitranslmed.adg3358.
10. Ferkingstad E, Sulem P, Atlason BA, Sveinbjornsson G, Magnusson MI, Styrmisdottir EL, Gunnarsdottir K, Helgason A, Oddsson A, Halldorsson BV, et al. Large-scale integration of the plasma proteome with genetics and disease. Nat Genet. 2021 Dec;53(12):1712-1721. doi: 10.1038/s41588-021-00978-w.
11. Aggarwal S, Karmakar A, Krishnakumar S, Paul U, Singh A, Banerjee N, Laha N, Roy Ball G, Srivastava S. Advances in Drug Discovery based on Genomics, Proteomics and Bioinformatics in Malaria. Curr Top Med Chem. 2023;23(7):551-578. doi: 10.2174/1568026623666230418114455.
12. Sadee W, Wang D, Hartmann K, Toland AE. Pharmacogenomics: Driving Personalized Medicine. Pharmacol Rev. 2023 Jul;75(4):789-814. doi: 10.1124/pharmrev.122.000810.
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


