A Comprehensive Guide to Quantitative Lipidomics: Methodologies, Workflows, and Applications
Lipids are no longer viewed merely as passive energy reservoirs but as a complex "language" governing cellular communication and systemic health. In the era of precision medicine, quantitative lipidomics has emerged as a transformative tool for unraveling the intricacies of lipid metabolism and its profound role in disease progression. By leveraging advanced LC-MS lipid analysis, researchers can now move beyond simple identification to achieve precise lipid quantification across thousands of molecular species. This ultimate guide explores how high-throughput lipid profiling empower lipid biomarker discovery and translate raw data into deep biological insights. Whether you are navigating drug development or fundamental life sciences, understanding the quantitative lipidome is essential for the next generation of biomedical breakthroughs.
1. Why Quantitative Lipidomics is Revolutionizing Biomedical Research
Lipids are fundamental architects and dynamic communicators of life. Far beyond their classic roles as passive structural components of membranes or inert energy stores, we now understand lipids as a vast, diverse class of molecules that actively govern cellular signaling, energy homeostasis, and stress responses. Specific lipid species, such as sphingolipids and phosphoinositides, act as crucial second messengers in signal transduction, while eicosanoids derived from polyunsaturated fatty acids are central mediators of inflammation. The composition of the cellular lipidome—encompassing thousands of unique molecular species—is exquisitely sensitive to genetic, dietary, and pathological perturbations, making it a rich, yet complex, source of biological insight into states of health and disease (Géhin et al., 2022). Therefore, comprehensively understanding a biological system necessitates a deep and systematic investigation of its lipids.
However, traditional lipid biochemistry approaches have long faced a fundamental limitation: they are largely targeted and qualitative. Methods like thin-layer chromatography or enzymatic assays can only provide information on broad lipid classes or a handful of predefined molecules, missing the vast molecular diversity within each class. These techniques cannot capture the system-wide, quantitative shifts in specific lipid molecular species (e.g., PC 34:1 vs. PC 36:2) that often underlie precise metabolic dysfunctions or early disease mechanisms. Quantitative lipidomics emerges as the essential solution to this bottleneck. By integrating advanced separation science with high-resolution mass spectrometry, it enables the simultaneous identification and precise quantification of hundreds to thousands of lipids in a single analysis. This shift from targeted, qualitative probing to untargeted, quantitative profiling allows researchers to move from asking “what lipids are there?” to answering the transformative question: “how do the absolute or relative levels of the entire lipid network change in response to a disease, drug, or genetic alteration?” This capability to measure dynamic flux is what positions quantitative lipidomics as an indispensable tool for unbiased biomarker discovery, elucidating complex disease etiologies, and evaluating drug efficacy and toxicity in modern biomedical research (Salihovic et al., 2023).
2. Key Considerations in High-Throughput Quantitative Lipidomics
The power of quantitative lipidomics lies not only in the technology itself but in the strategic decisions made at the outset of a study. Two fundamental choices—selecting the appropriate analytical platform and defining the experimental strategy—directly determine the scope, depth, and applicability of the results.
2.1 LC-MS vs. GC-MS: Choosing the Best Platform for Lipid Analysis
The cornerstone of modern lipidomics is mass spectrometry (MS), but the choice of front-end separation technique is critical. Today, liquid chromatography-mass spectrometry (LC-MS) is the undisputed workhorse for global lipid profiling. Its dominance stems from its exceptional versatility and compatibility with the vast majority of lipid classes. LC-MS gently separates lipids in a liquid phase based on properties like polarity (using reverse-phase columns like C18) or headgroup (using hydrophilic interaction chromatography, HILIC), preserving intact molecular ions for analysis. This makes it ideal for thermally labile and high-molecular-weight lipids such as phospholipids, sphingolipids, and triacylglycerols. The high sensitivity and broad dynamic range of modern tandem mass spectrometers (MS/MS) allow for the detection and identification of hundreds to thousands of lipid species from a single, small-volume sample in an untargeted lipid profiling experiment (Cai et al., 2024).
In contrast, gas chromatography-mass spectrometry (GC-MS) plays a complementary, targeted role. It excels at the precise quantification of volatile or volatilizable lipids. This makes it the gold standard for analyzing short-chain fatty acids (SCFAs) from gut microbiota or specific fatty acid methyl esters (FAMEs) following derivatization. GC-MS offers superior chromatographic resolution for these small molecules and leverages extensive, well-established spectral libraries. However, its requirement for high-temperature vaporization limits its application to intact, large, or polar lipid molecules. Thus, while LC-MS provides the panoramic view, GC-MS offers unmatched precision for specific, predefined metabolic targets
2.2 Untargeted vs. Targeted Lipidomics: Selecting the Right Strategy
The analytical goal dictates the strategy, broadly categorized into two paradigms: untargeted and targeted lipidomics.
Untargeted Lipidomics is the exploratory powerhouse. Operating on a “measure everything possible” principle, it is a hypothesis-generating approach designed for biomarker discovery and novel pathway identification. By performing a comprehensive, unbiased scan of the lipidome, it captures both expected and unexpected changes, creating a global snapshot of lipid metabolism. This strategy is the essential first step in most research pipelines, revealing which lipid families and individual species are differentially regulated in response to a disease state, genetic modification, or drug treatment.
Once key lipids of interest are pinpointed through untargeted screening, Targeted Lipidomics takes over for validation and precise measurement. This strategy uses highly optimized methods, such as multiple reaction monitoring (MRM) on triple quadrupole MS, to focus with extreme sensitivity and specificity on a predefined list of lipids. It is designed for accuracy, precision, and high-throughput quantification across large sample cohorts. Targeted analysis answers confirmatory questions: “Exactly how much of this specific biomarker lipid is present in each of these 500 patient samples?” It is the cornerstone of translational research and clinical assay development.
3. The Quantitative Lipidomics Workflow: From Sample to Biological Insight
A successful quantitative lipidomics study is built on a meticulously executed, multi-step workflow. Each stage, from the initial handling of your precious sample to the generation of raw instrument data, is critical for ensuring the final biological conclusions are both reliable and meaningful.
3.1 Sample Preparation and Lipid Extraction
In lipidomics, the principle of “garbage in, garbage out” is paramount. The integrity of your final data is fundamentally determined by the initial handling and processing of your samples. Consistent, meticulous preparation is essential to minimize pre-analytical variation and ensure the measured lipid profile reflects true biology, not artifacts. Common sample types—plasma/serum, tissues (e.g., liver, brain), and cultured cells—each require tailored protocols, but universal rules apply: maintain a cold chain using pre-chilled solvents and equipment, process samples rapidly to quench enzymatic activity, and strictly avoid repeated freeze-thaw cycles. For oxidation-prone lipids, adding antioxidants to the lysis buffer is standard practice.
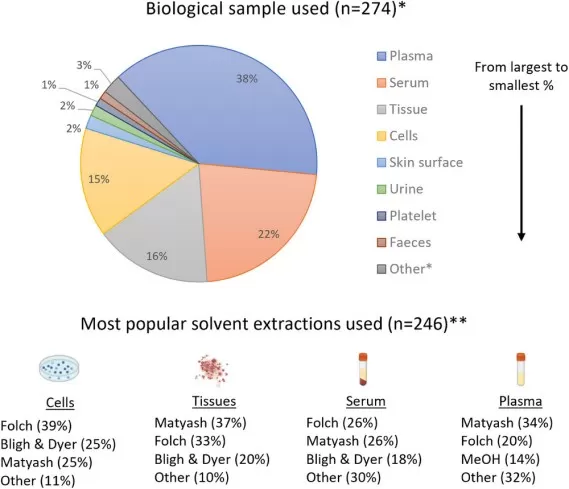
Distribution of Primary Sample Types and Extraction Methods in Clinical Lipidomics Research (Bibliometric Analysis, 2022)
Data and Image reproduced from Géhin et al., 2022, Analytical science advances, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
Following stabilization, the goal of lipid extraction is to efficiently and selectively isolate the lipidome from proteins, sugars, and salts. The choice of extraction method balances efficiency, safety, and compatibility with downstream LC-MS analysis. Several well-established liquid-liquid extraction methods are employed, each with its own advantages: The MTBE (methyl-tert-butyl ether) method is widely adopted in modern high-throughput labs due to its lower toxicity, clean phase separation, and high recovery rates. The classical Folch (chloroform:methanol) and Bligh & Dyer methods remain gold standards for their robust and broad-spectrum recovery. The choice often depends on sample type, safety considerations, and downstream compatibility with the LC-MS system. The MTBE method has gained significant popularity due to its superior safety profile and robust performance.
Comparison of Three Classic Lipid Extraction Methods: Folch, Bligh & Dyer, and MTBE
|
Method |
Principle & Composition |
Key Advantages |
Ideal For |
|
Folch |
Chloroform: Methanol (2:1 v/v) |
Considered the “gold standard”; excellent recovery of most lipid classes. |
Broad-spectrum extraction, especially for phospholipids and neutral lipids. |
|
Bligh & Dyer |
Chloroform: Methanol: Water (1:2:0.8 v/v) |
Adapted for watery samples; effective for tissues with high water content. |
Direct extraction from moist tissues or biological fluids. |
|
MTBE (Methyl-tert-butyl ether) |
MTBE: Methanol: Water (10:3:2.5 v/v) |
Less toxic than chloroform; cleaner phase separation; high efficiency. |
Modern high-throughput labs; excellent for a wide range of sample types. |
3.2 LC-MS Data Acquisition for Lipid Profiling
The purified lipid extract is then introduced into the liquid chromatography-mass spectrometry (LC-MS) system, which acts as a high-resolution molecular scanner to separate, identify, and quantify individual lipid species.
1) Chromatographic Separation: Reducing Complexity
Direct infusion of a total lipid extract leads to ion suppression and uninterpretable signal overlap. Liquid chromatography (LC) solves this by separating lipids in time based on their physicochemical properties. The choice of chromatographic column is strategic:
- Reversed-Phase (e.g., C18, C30): The most common approach. Lipids are separated based on the hydrophobicity of their fatty acyl chains. C18 columns provide excellent resolution for most molecular species. C30 columns, with denser packing, offer enhanced separation for challenging isomers like triglycerides or sphingolipids with similar chain lengths.
- Hydrophilic Interaction Liquid Chromatography (HILIC): This mode separates lipids based on the polarity of their head groups. It is invaluable for separating lipid classes (e.g., PC from PE from PI) that co-elute on reversed-phase columns, providing complementary information.
2) Mass Spectrometric Detection: The Core of Analysis
Following separation, lipids are ionized (typically via electrospray ionization) and analyzed by the mass spectrometer. The MS performs two key functions: 1) Identification, by measuring the precise mass-to-charge ratio (m/z) of the intact ion and generating a characteristic fragmentation pattern (MS/MS spectrum) for database matching; and 2) Quantification, by recording the signal intensity of specific ions. The acquisition mode is strategically chosen: Data-Dependent Acquisition (DDA) is ideal for untargeted discovery, automatically fragmenting abundant ions to identify unknowns, while Multiple Reaction Monitoring (MRM) is used for targeted quantification, offering superior sensitivity and precision for validating predefined lipids.
3) Batch Quality Control: Ensuring Instrument Stability
A typical lipidomics run involves analyzing dozens to hundreds of samples over hours or days. To monitor and ensure consistent instrument performance, Quality Control (QC) samples—prepared from a pooled aliquot of all study samples—are injected at regular intervals throughout the sequence. By tracking the retention time, peak shape, and intensity of key lipids in these QCs, scientists can detect and correct for any instrumental drift, guaranteeing data consistency and reliability from the first injection to the last.
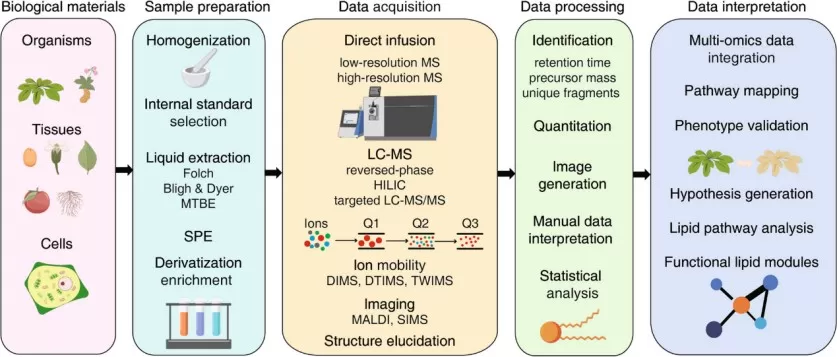
The lipidomics workflow
Image reproduced from Liu et al., 2023, Current opinion in chemical biology, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
3.3 Lipid Identification and Quantification
This stage transforms raw mass spectral data into a structured, analyzable dataset. Bioinformatics software first performs peak picking and alignment, detecting chromatographic features and matching them across all samples to create a consistent feature list. Lipid identification is then achieved by matching the precise mass and fragmentation patterns (MS/MS spectra) of these features against curated public databases like LIPID MAPS, often augmented by in-house spectral libraries for higher confidence and coverage.
In discovery lipidomics, identifications are typically reported at the molecular species level (e.g., PC 34:1). Finally, quantitation is performed by normalizing the peak intensity of each identified lipid to class-specific internal standards added during sample preparation. This process generates the core data matrix—lipids in rows, samples in columns, with values representing relative abundances. For untargeted, high-throughput studies, relative quantitation (measuring fold-changes) is the mainstream approach, as it efficiently identifies significant alterations without the prohibitive cost and complexity of absolute quantification for thousands of lipids.
3.4 Data Analysis and Biological Interpretation
The final step extracts biological meaning from the data matrix through rigorous statistical and bioinformatic analysis. The process begins with data quality control and normalization to remove technical noise. Multivariate statistics like PCA provide an overview of sample grouping and outliers, while univariate tests identify individual lipids with significant abundance changes between conditions.
To move beyond a list of differences, pathway and enrichment analysis maps these altered lipids onto known metabolic networks (e.g., glycerophospholipid metabolism), revealing coherent biological themes. Furthermore, lipid-specific analyses offer unique insights: examining changes at the lipid class level can indicate shifts in enzymatic pathways, and calculating indices like the Double Bond Index provides a snapshot of overall lipid saturation related to membrane fluidity or oxidative stress. This layered interpretation transforms complex data into a actionable biological narrative, highlighting key mechanisms and candidate biomarkers for further validation.
4. Key Applications of Quantitative Lipidomics in Biological Research
Quantitative lipidomics serves as a powerful discovery engine across biomedical and life sciences, translating complex lipid signatures into actionable insights for diagnosis, mechanistic understanding, and product development.
4.1 Lipid Biomarker Discovery for Disease Diagnosis and Prognosis
Quantitative lipidomics is pivotal for biomarker discovery, systematically comparing lipid profiles to identify molecular signatures with diagnostic or prognostic power. Its high sensitivity detects subtle metabolic shifts often preceding clinical symptoms. A prime example is its application in stratifying infectious diseases. A recent untargeted lipidomics study of COVID-19 patient plasma revealed a panel of lipids, including specific ceramides and sphingosine-1-phosphate, whose levels robustly distinguished severe from mild cases with superior accuracy compared to standard clinical markers (Sun et al., 2024). This demonstrates its potential to uncover high-performance predictive biomarkers for patient triage and management.
4.2 Lipid Metabolism in Disease Mechanisms
Beyond listing changes, lipidomics deciphers disease mechanisms by mapping alterations to specific metabolic pathways, revealing how pathologies rewire lipid metabolism. This is crucial in complex diseases like neurodegeneration. Research synthesizing lipidomics findings has shown that distinct alterations in brain glycosphingolipids are closely tied to key pathological processes in various dementias, such as neuroinflammation and protein aggregation (Sarbu et al., 2025). These findings position specific lipid species not only as biomarkers but as active players and potential therapeutic targets in the disease cascade.
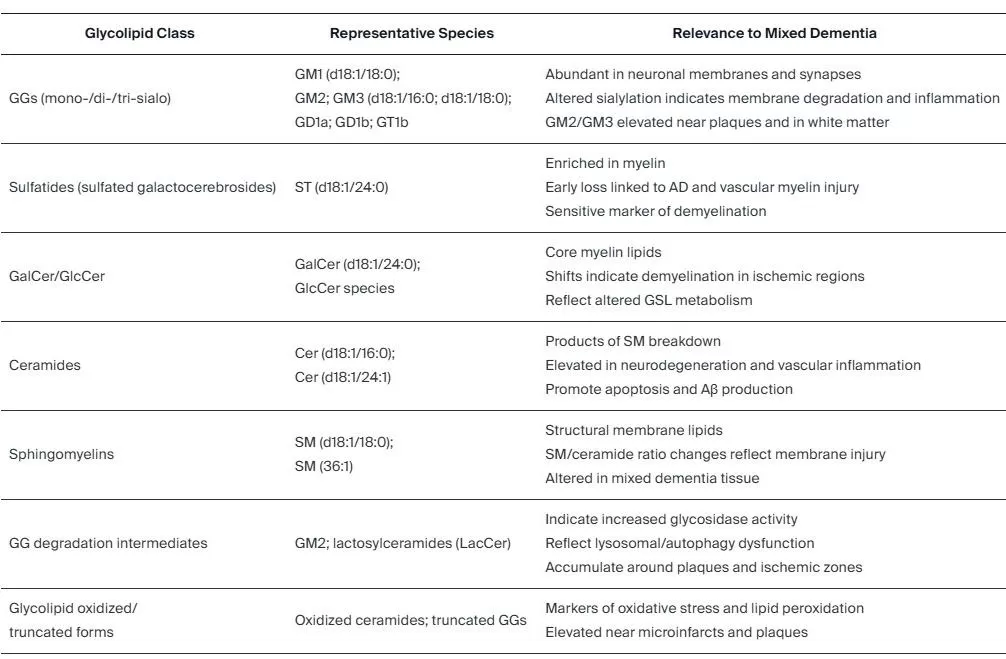
Glycolipid classes relevant to mixed dementia discovered by MS-based lipidomics approaches.
Table reproduced from Sarbu et al., 2025, Biomedicines, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
4.3 Lipidomics in Drug Development and Toxicology Assessment
In drug development, lipidomics provides a unique window into drug efficacy and safety by profiling global lipidomic responses to therapy. It can identify predictive biomarkers of response and uncover lipid-related adverse effects. For instance, in non-small cell lung cancer, a pre-treatment plasma lipid signature (including linoleic acid and specific phospholipids) was found to predict patient response to first-line immunotherapy combined with chemotherapy (Yu et al., 2025). This approach aids in personalizing treatment, while mechanistic insights into key lipids can inform the development of novel combination strategies.
4.4 Exploring Lipidomics in Nutrition and Agricultural Sciences
In food and agricultural sciences, lipidomics precisely tracks how genetics, diet, and processing affect lipid composition, guiding efforts to improve nutritional quality and crop traits. It acts as a detailed analytical tool for quality control. A relevant study used lipidomics to monitor the physical refining of safflower and flaxseed oils, revealing that the deacidification step significantly alters the levels of nutritionally critical polyunsaturated fatty acids and phospholipids (Team Research, 2025). This enables the optimization of processing conditions to better preserve the health-promoting components of food oils.
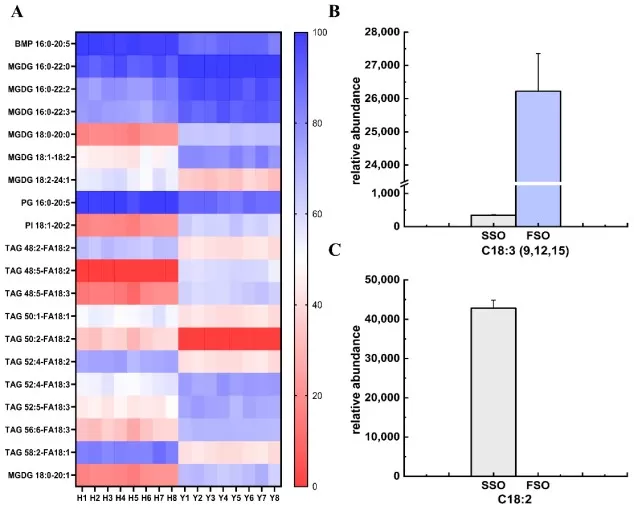
The results of heatmaps of lipids and screenings of FA with significant differences in safflower seed oil (SSO) and flaxseed oil (FSO).
Image reproduced from Yang et al., 2025, Foods, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
5. Why Choose MetwareBio for Quantitative Lipidomics Service?
Navigating the complex world of lipidomics requires more than just advanced instrumentation; it demands a partner with proven expertise, robust methodology, and a commitment to delivering biologically meaningful data. At MetwareBio, we have engineered our quantitative lipidomics service to meet the highest standards of precision, depth, and reliability, providing researchers with a trusted foundation for groundbreaking discoveries.
5.1 Comprehensive Coverage: 4000+ Lipids Across 6 Major Lipid Classes
Achieving a complete picture of the lipidome is the first step to meaningful insight. Our service is powered by an extensive in-house library and advanced analytical platforms, enabling the simultaneous identification and relative quantification of over 4,000 lipid molecules. This comprehensive profiling spans all six major lipid classes—fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterol lipids, and prenol lipids. Whether you are investigating energy metabolism, membrane dynamics, or inflammatory signaling, our broad coverage ensures that no key player in your biological system goes unnoticed, turning a snapshot into a detailed metabolic map.
The lipid database of Metwarebio’s Quantitative Lipidomics Service
|
Class I |
Class II |
Number |
|
Fatty acyls(FA) |
CAR, FFA, Eicosanoid, FAHFA |
270 |
|
Glycerolipids(GL) |
DG, DG-O, MG, TG, TG-O, MGDG, DGDG |
1015 |
|
Glycerophospholipids(GP) |
LPC, LPC-O, LPE, LPE-P, LPG, LPS, PC, PC-O, PE, PE-P, PE-O, PG, PS, LPI, PI, LPA, PA, PMeOH, BMP, HMBP, LNAPE |
1800 |
|
Sphingolipids(SL) |
SPH, CerP, HexCer, SM, Cer, Cert |
828 |
|
Sterol lipids(ST) |
Cho, CE, BA, CASE |
122 |
|
Prenol lipids(PR) |
CoQ |
3 |
|
Total |
|
4000+ |
5.2 Highly Accurate Quantification with Internal Standards and MRM Mode
Precision is non-negotiable in quantitative science. We ensure data accuracy and reproducibility through a rigorous, multi-layered quality control system. Every analysis incorporates a comprehensive panel of isotope-labeled internal standards for robust calibration and employing MRM on QTRAP systems for highly selective detection. This integrated approach enables reliable absolute quantification of lipids with sensitivity at the nmol level, ensuring the accuracy and reproducibility required for demanding research. From stringent sample processing SOPs to the inclusion of quality control samples in every batch, our entire pipeline is designed to deliver the reliable, publication-ready quantitative data that your research demands.
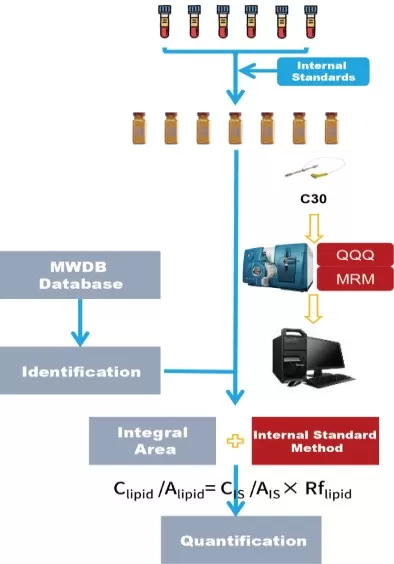
The analytical workflow of Metwarebio’s Quantitative Lipidomics Service
5.3 Advanced Technical Support and Tailored Solutions for Your Research
We understand that every research project is unique. That’s why our service extends beyond standardized reporting to offer personalized expert support. Our team of experienced scientists works with you from project design to data interpretation, helping to tailor the analysis to your specific goals—whether you need to focus on a specific pathway (like sphingolipid metabolism in neurological studies), validate biomarkers from a discovery screen, or integrate lipidomics data with other omics layers. We provide clear, insightful biological interpretation of your results, helping you translate complex data into a compelling scientific narrative for your next grant, publication, or therapeutic development milestone.
Reference
1. Géhin, C., Fowler, S. J., & Trivedi, D. K. (2023). Chewing the fat: How lipidomics is changing our understanding of human health and disease in 2022. Analytical science advances, 4(3-4), 104–131. https://doi.org/10.1002/ansa.202300009
2. Salihovic, S., Lamichane, S., Hyötyläinen, T., & Orešič, M. (2023). Recent advances towards mass spectrometry-based clinical lipidomics. Current opinion in chemical biology, 76, 102370. https://doi.org/10.1016/j.cbpa.2023.102370
3. Liu, H. Z., Li, Y. K., Chen, Y. L., Zhou, Y., Sahu, S. K., Liu, N., Wu, H., Shui, G., Chen, Q., & Yao, N. (2024). Exploring the plant lipidome: techniques, challenges, and prospects. Advanced biotechnology, 2(1), 11. https://doi.org/10.1007/s44307-024-00017-9
4. Sun, J., Peters, M., Yu, L. R., Vijay, V., Bidarimath, M., Agrawal, M., Flores-Torres, A. S., Green, A. M., Burkhart, K., Oliphant, J., Smallwood, H. S., & Beger, R. D. (2024). Untargeted metabolomics and lipidomics in COVID-19 patient plasma reveals disease severity biomarkers. Metabolomics : Official journal of the Metabolomic Society, 21(1), 3. https://doi.org/10.1007/s11306-024-02195-y
5. Sarbu, M., Ica, R., Biricioiu, M. R., Dehelean, L., & Zamfir, A. D. (2025). Glycosphingolipids in Dementia: Insights from Mass Spectrometry and Systems Biology Approaches. Biomedicines, 13(12), 2854. https://doi.org/10.3390/biomedicines13122854
6. Yu, J., Xu, H., Xiong, F., Liu, X., Lingfei, M., Gao, H., & Li, Y. (2025). Lipidomics reveals biomarkers of the efficacy of first-line ICI therapy combined with chemotherapy in NSCLC. Journal of translational medicine, 23(1), 638. https://doi.org/10.1186/s12967-025-06542-y
7. Yang, J., Zhao, H., Wu, F., Wang, Z., Yuan, L., Qiu, Y., Wang, L., & Zhu, M. (2025). Lipidomics Approach Reveals the Effects of Physical Refining Processes on the Characteristic Fatty Acids and Physicochemical Indexes of Safflower Seed Oil and Flaxseed Oil. Foods (Basel, Switzerland), 14(16), 2845. https://doi.org/10.3390/foods14162845
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


