What Are Metabolites? Definition, Biological Significance, and Analytical Approaches
Living systems are defined not only by their genes or proteins, but by the countless small molecules that constantly flow through cells, tissues, and biofluids. These molecules—collectively known as metabolites—represent the most immediate and dynamic readout of biological activity. Whether a cell is growing, differentiating, responding to stress, or developing disease, these processes are ultimately reflected in its metabolic state.
In recent years, advances in metabolomics have made it possible to measure hundreds to thousands of metabolites simultaneously, providing unprecedented insight into the metabolome and its regulation. However, for many researchers who are new to the field, fundamental questions remain: What exactly are metabolites? How do they relate to metabolism and metabolic pathways? And why are they so central to modern biological and biomedical research?
This article provides a systematic introduction to metabolites, from their biochemical definition to their role in physiology and disease, and explains how metabolomics enables comprehensive metabolite profiling across diverse biological systems.
What Are Metabolites? A Biochemical Definition
In biochemistry, a metabolite is typically defined as a small molecule that participates in, or is produced by, metabolic reactions within a living organism. Unlike macromolecules such as DNA, RNA, or proteins, metabolites are generally low–molecular weight compounds, often below 1,500 Da, and include a wide chemical diversity.
Metabolites can be broadly classified into two categories based on their biological roles. Primary metabolites are directly involved in essential cellular functions such as energy production, growth, and maintenance. These include amino acids, sugars, organic acids, nucleotides, and lipids that form the core of central metabolism. Secondary metabolites, sometimes referred to as specialized metabolites, are not strictly required for survival under laboratory conditions but play important roles in adaptation, signaling, and interaction with the environment. Examples include plant flavonoids, alkaloids, terpenoids, and microbial antibiotics.
Importantly, metabolites are not static entities. Their concentrations can change rapidly in response to nutrient availability, environmental stimuli, genetic perturbations, or disease processes. This dynamic nature makes metabolites uniquely informative compared with genes or proteins, whose levels often change more slowly or indirectly. From a systems biology perspective, metabolites represent the functional output of gene expression, enzyme activity, and regulatory networks, providing a direct snapshot of cellular physiology.
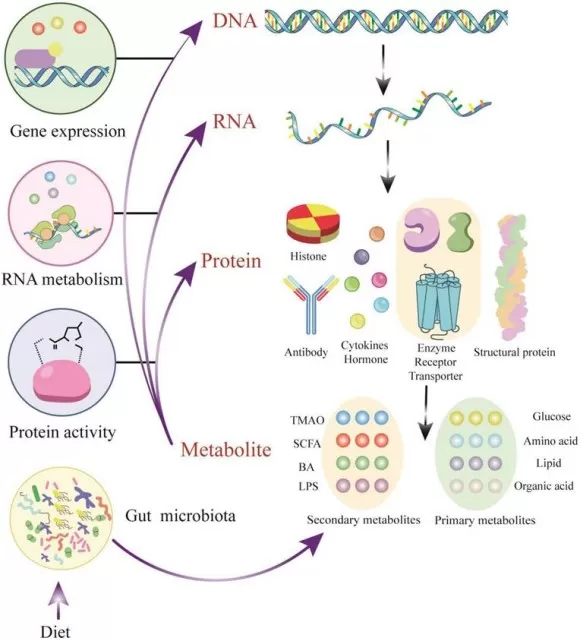
Metabolites - active modulators of gene and protein activity
Image reproduced from Wang et al., 2023, International Journal of Biological Sciences, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
From Metabolism to the Metabolome: Understanding Metabolic Organization
To fully appreciate the importance of metabolites, it is helpful to place them within the broader framework of metabolism. Metabolism encompasses all chemical reactions that occur in a biological system and is traditionally divided into catabolism, which breaks down molecules to generate energy and building blocks, and anabolism, which uses these resources to synthesize complex biomolecules.
These reactions are organized into interconnected metabolic pathways, such as glycolysis, the tricarboxylic acid (TCA) cycle, amino acid metabolism, lipid metabolism, and nucleotide biosynthesis. Each pathway consists of sequential enzymatic steps, transforming one metabolite into another with remarkable specificity and regulation.
The metabolome refers to the complete set of metabolites present in a cell, tissue, organ, or biofluid at a given time and under defined conditions. Unlike the genome, which is largely stable, the metabolome is highly context-dependent. It varies across cell types, developmental stages, physiological states, and environmental conditions.
This context sensitivity is precisely what makes metabolite analysis so powerful. While genomic and transcriptomic data describe what could happen in a biological system, the metabolome reflects what is actually happening at the biochemical level.
Why Metabolites Matter: Biological and Clinical Significance
Metabolites as the Functional Core of Cellular Biology
At the most fundamental level, metabolites are indispensable to cellular life. They provide the chemical energy required to sustain biological processes, participate in redox reactions that maintain cellular homeostasis, and regulate osmotic balance within cells and tissues. Many metabolites act as substrates, intermediates, or cofactors for enzymatic reactions, thereby enabling the continuous flow of metabolism that supports growth, maintenance, and adaptation.
Beyond these classical biochemical roles, metabolites are increasingly recognized as active regulators of cellular function rather than passive byproducts of metabolism. Fluctuations in metabolite availability can directly influence enzyme activity and metabolic flux, shaping how cells allocate resources in response to internal demands and external conditions. In this sense, metabolites represent the functional currency of biological systems.
Metabolites as Signaling Molecules and Regulatory Factors
In addition to their metabolic roles, many metabolites function as signaling molecules that modulate gene expression, epigenetic states, and intercellular communication. Certain metabolites can act as ligands for nuclear receptors or other signaling proteins, linking metabolic status to transcriptional regulation. Others influence chromatin structure and epigenetic modifications by serving as substrates or inhibitors of enzymes involved in DNA and histone modification.
Through these mechanisms, metabolites help coordinate cellular behavior with nutrient availability, energy status, and environmental cues. This metabolic regulation extends beyond individual cells, contributing to tissue-level coordination and organism-wide physiological responses. As a result, changes in metabolite levels can have far-reaching effects that go well beyond metabolism itself.
Metabolites in Physiology and Disease: Early Indicators of Biological Change
In both normal physiology and disease, alterations in metabolite levels often occur earlier than detectable changes at the protein or phenotypic level. Because metabolites respond rapidly to perturbations, they provide a sensitive window into emerging biological changes. For example, disruptions in glucose and lipid metabolism are hallmark features of metabolic disorders such as diabetes and obesity, reflecting impaired energy homeostasis long before structural damage becomes apparent.
Similarly, rewiring of amino acid, nucleotide, and lipid metabolism is commonly observed in cancer, immune activation, and neurodegenerative diseases. These metabolic alterations support processes such as rapid cell proliferation, inflammation, or neuronal dysfunction. By capturing such changes, metabolomics enables researchers to study disease mechanisms at a stage when intervention may be most effective.
Metabolites as Biomarkers and Mediators of Host–Microbiome Interactions
Because metabolites integrate genetic background with environmental influences—including diet, lifestyle, and microbial activity—they are particularly valuable as biomarkers. Metabolite signatures measured in plasma, serum, urine, or cerebrospinal fluid can reflect disease risk, progression, and response to therapy, offering practical advantages for translational and clinical research.
Metabolites also serve as the biochemical interface between the host and the microbiome. Microbial-derived metabolites can enter host circulation and influence host metabolism, immune responses, and even neurological function. This bidirectional metabolic communication has positioned metabolomics as a central tool in microbiome research and in integrated multi-omics studies aimed at understanding complex host–environment interactions.
What Is Metabolomics? Measuring Metabolites at Scale
Metabolomics is the systematic study of metabolites and metabolic pathways using high-throughput analytical technologies. Its goal is to comprehensively profile the metabolome and quantify changes in metabolite abundance across different biological conditions.
Unlike targeted biochemical assays that focus on a small number of predefined compounds, metabolomics can capture a broad spectrum of metabolites simultaneously. This global or semi-global view is critical for discovering unexpected metabolic alterations and generating new biological hypotheses.
Modern metabolomics relies primarily on mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy. Among these, LC-MS–based metabolomics has become the most widely used approach due to its high sensitivity, broad metabolite coverage, and compatibility with diverse sample types.
Depending on study objectives, metabolomics experiments can be designed as untargeted metabolomics, which aims to profile as many metabolites as possible without prior selection, or targeted metabolomics, which focuses on a defined panel of metabolites with high quantitative accuracy. Each approach has distinct advantages, and the choice often depends on whether the goal is discovery-driven research or hypothesis-driven validation.
Regardless of the analytical strategy, metabolomics typically involves several key steps, including sample collection and preparation, metabolite extraction, data acquisition, metabolomics data analysis, metabolite identification, and metabolic pathway analysis. Careful experimental design and standardized workflows are essential to ensure data quality and biological interpretability.
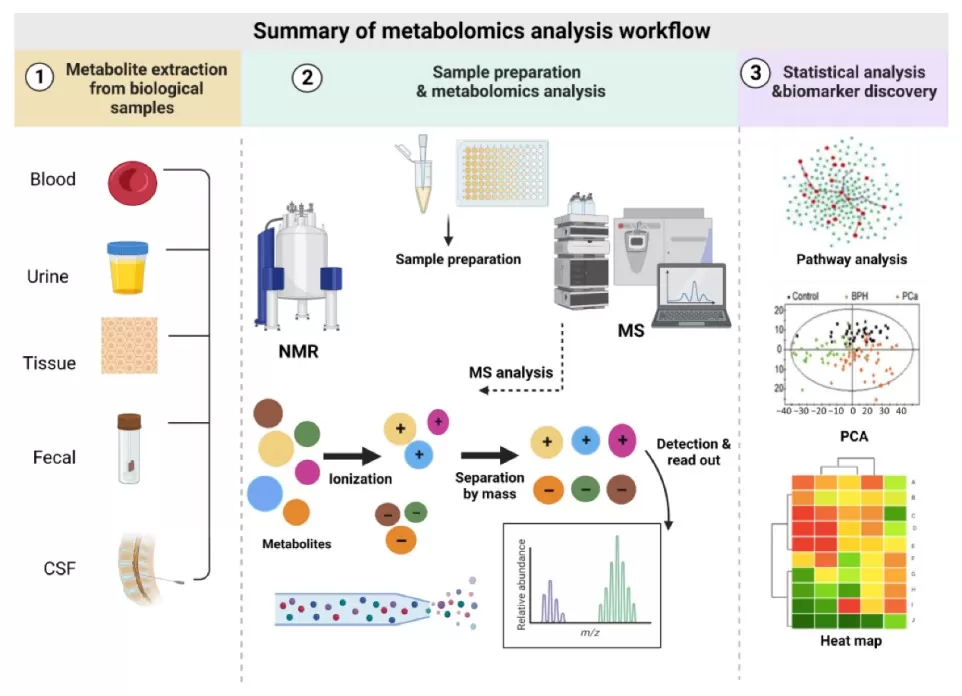
Summary of the metabolomics analysis workflow
Image reproduced from Al-Sulaiti et al., 2023, Metabolites, licensed under CC BY 4.0.
Metabolite Profiling and Metabolic Pathway Analysis
At the heart of metabolomics lies metabolite profiling—the measurement of relative or absolute concentrations of metabolites across samples. By comparing metabolite profiles between experimental groups, researchers can identify significantly altered metabolites and infer affected metabolic pathways.
Metabolic pathway analysis integrates metabolomics data with curated biochemical knowledge to reveal coordinated changes in metabolism. Rather than interpreting individual metabolites in isolation, pathway-level analysis provides a systems-level view of metabolic reprogramming. This approach is particularly valuable in complex biological contexts, where subtle but coordinated changes across multiple metabolites may reflect underlying regulatory mechanisms. Tools such as pathway enrichment analysis and network-based visualization help translate metabolomics results into biologically meaningful insights.
Importantly, metabolomics data are often most powerful when combined with other omics layers, such as genomics, transcriptomics, or proteomics. In integrated multi-omics studies, metabolites serve as functional anchors that connect molecular regulation with phenotype.
Looking Ahead: Metabolites as a Window into Biological Function
As analytical technologies and computational methods continue to advance, metabolomics is becoming an increasingly central component of life science research. From basic studies of cellular metabolism to translational applications in precision medicine, the ability to systematically analyze metabolites is reshaping how researchers understand biological systems.
Metabolites offer a unique perspective: they are chemically diverse, highly dynamic, and directly linked to phenotype. By studying the metabolome, scientists can move beyond static descriptions of biological potential and toward a functional understanding of metabolic activity.
For researchers seeking to explore metabolic mechanisms, identify biomarkers, or integrate metabolism into multi-omics studies, metabolomics provides a powerful and versatile toolkit. A solid understanding of what metabolites are, how they function, and how they can be measured is therefore an essential foundation for modern biological and biomedical research.
Frequently Asked Questions About Metabolites and Metabolomics
1. What is a metabolite?
A metabolite is a small molecule that is involved in metabolic reactions within a living system. Metabolites can act as substrates, intermediates, or end products of metabolism, and they directly reflect the biochemical activity of cells, tissues, or organisms at a given time. Because metabolite levels can change rapidly in response to genetic, environmental, or physiological factors, they provide a highly sensitive readout of biological function.
2. What is the difference between metabolites and the metabolome?
Metabolites are individual small molecules, such as amino acids, sugars, lipids, or organic acids. The metabolome, in contrast, refers to the complete set of metabolites present in a biological system under specific conditions. While individual metabolites offer limited information on their own, analysis of the metabolome enables a global view of metabolic activity and pathway-level regulation.
3. What is metabolomics and what does it measure?
Metabolomics is the systematic study of metabolites using high-throughput analytical technologies such as mass spectrometry or nuclear magnetic resonance spectroscopy. It measures changes in metabolite abundance across biological samples to characterize metabolic pathways, biological responses, and disease-associated metabolic alterations. As a result, metabolomics provides functional insights that complement other omics approaches.
4. Why are metabolites considered close to phenotype?
Metabolites are often described as being “closest to phenotype” because they represent the final outcome of gene expression, protein activity, and environmental influences. Unlike genomic or transcriptomic data, which reflect biological potential, metabolites capture real-time biochemical activity and therefore provide a direct link between molecular regulation and observable phenotypes.
5. What types of metabolites are typically analyzed in metabolomics studies?
Metabolomics studies commonly analyze metabolites involved in central carbon metabolism, amino acid metabolism, lipid metabolism, nucleotide metabolism, and redox processes. Depending on the biological system, specialized or secondary metabolites—such as plant natural products or microbial metabolites—may also be included. The exact metabolite coverage depends on the analytical platform and study design.
6. What is the difference between targeted and untargeted metabolomics?
Untargeted metabolomics aims to detect as many metabolites as possible without prior selection, making it well suited for discovery-driven research. Targeted metabolomics, by contrast, focuses on a predefined set of metabolites and offers higher quantitative accuracy and sensitivity. The choice between these approaches depends on the scientific question and experimental objectives.
7. What biological samples can be used for metabolomics analysis?
A wide range of biological samples can be analyzed using metabolomics, including plasma, serum, urine, tissues, cultured cells, cerebrospinal fluid, and microbial or plant samples. Each sample type requires appropriate collection, storage, and preparation procedures to preserve metabolite integrity and ensure reliable metabolic profiling.
8. How are metabolites identified in metabolomics studies?
Metabolite identification typically relies on accurate mass measurements, chromatographic retention time, and MS/MS fragmentation patterns. These data are compared against spectral libraries and reference standards. Because many metabolites share similar chemical properties, confident identification often requires multiple lines of evidence and careful data interpretation.
9. What is metabolic pathway analysis and why is it important?
Metabolic pathway analysis integrates metabolomics data with curated biochemical pathway information to identify coordinated changes in metabolism. By focusing on pathways rather than individual metabolites alone, researchers can gain deeper insight into biological mechanisms and regulatory processes underlying observed metabolic alterations.
10. How does metabolomics complement other omics approaches?
Metabolomics complements genomics, transcriptomics, and proteomics by providing a functional biochemical readout of biological systems. While other omics layers describe genetic potential or regulatory changes, metabolomics captures the metabolic consequences of those changes, making it an essential component of integrated multi-omics studies.
Reference
1. Wang Q, Yesitayi G, Liu B, Siti D, Ainiwan M, Aizitiaili A, Ma X. Targeting metabolism in aortic aneurysm and dissection: from basic research to clinical applications. Int J Biol Sci. 2023 Jul 31;19(12):3869-3891. doi: 10.7150/ijbs.85467. PMID: 37564200; PMCID: PMC10411465.
2. Al-Sulaiti H, Almaliti J, Naman CB, Al Thani AA, Yassine HM. Metabolomics Approaches for the Diagnosis, Treatment, and Better Disease Management of Viral Infections. Metabolites. 2023 Aug 15;13(8):948. doi: 10.3390/metabo13080948. PMID: 37623891; PMCID: PMC10456346.
Read more
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


