MTBE vs. Chloroform–Methanol Lipid Extraction: Which Method Fits Your Lipidomics Workflow?
In lipidomics, lipid extraction is the first and often most influential step affecting downstream data quality. Extraction performance determines (i) overall recovery, (ii) class‑dependent bias (e.g., preferential loss of highly polar lipids), (iii) the extent of matrix carryover (proteins, salts, detergents) that can suppress ionization, and (iv) technical reproducibility across batches. Consequently, a well‑controlled extraction workflow is essential for reliable LC–MS lipidomics and shotgun lipidomics.
Two families of methods are most widely used in routine laboratories: the long‑established chloroform–methanol protocols (commonly represented by the Folch and Bligh & Dyer methods) and the newer MTBE (methyl tert‑butyl ether) protocol [1-2]. Researchers often ask which method best fits their sample type and study objectives, and how the methods differ in safety, efficiency, and practicality. This report provides a structured comparison, highlights common pitfalls, and proposes actionable best practices to improve extraction robustness.
1. Core Principle: Biphasic Partitioning with Different Phase Geometry
Both approaches rely on solvent polarity and phase partitioning: lipids preferentially dissolve in a non‑polar organic phase, whereas proteins, salts, and other hydrophilic components remain in the aqueous phase or at the interphase. The key operational difference is the density of the organic solvent, which determines whether the lipid‑rich phase forms at the bottom or the top. This phase geometry strongly influences ease of handling and risk of interphase contamination.
1.1 Chloroform–Methanol Extraction (Folch / Bligh & Dyer)
Chloroform acts as the primary hydrophobic solvent that efficiently solubilizes a wide range of lipid classes. Methanol serves as a polar co‑solvent that improves miscibility during extraction, promotes protein precipitation, and disrupts membrane structures to release lipids. After water addition, a biphasic chloroform–methanol–water system forms. Because chloroform is dense (≈1.48 g/cm³), the lipid‑enriched organic phase is the lower layer, while the aqueous phase remains on top and insoluble material concentrates at the interphase [1].
In practice, the Folch method is often applied to lipid‑rich samples, whereas the Bligh & Dyer method adjusts solvent ratios and is commonly preferred for high‑water‑content matrices such as plasma and tissue homogenates.
1.2 MTBE Extraction
The MTBE method uses MTBE as the hydrophobic solvent and methanol as a co‑solvent to form a biphasic system after water addition. Because MTBE has low density (≈0.74 g/cm³), the lipid‑rich organic phase forms the upper layer. The aqueous phase typically occupies the middle, and precipitated proteins/cellular debris compact as a pellet at the bottom. This configuration generally simplifies organic‑phase collection and reduces the likelihood of aspirating interphase material [2].
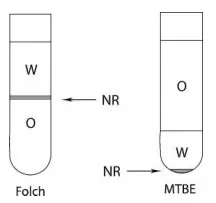
Lipid extraction for high throughput lipidomics [2]
2. Simplified Workflow Comparison: Practical Convenience
Both methods follow the same high‑level workflow (solvent addition → mixing/extraction → phase separation → organic‑phase collection → drying → reconstitution). However, the location of the organic phase results in a meaningful ergonomic difference: MTBE enables straightforward top‑phase collection, whereas chloroform–methanol requires careful bottom‑phase aspiration through the aqueous layer and interphase [3].
|
Step |
Chloroform–MeOH (Folch/Bligh & Dyer) |
MTBE–MeOH |
|
Extraction |
MeOH + CHCl₃; mix ~1 h (RT) |
MeOH + MTBE; mix ~1 h (RT) |
|
Phase split |
Add H₂O; organic = bottom |
Add H₂O; organic = top |
|
Collection |
Aspirate bottom; avoid interphase |
Aspirate top; avoid aqueous |
|
Finish |
Combine → dry → reconstitute |
Combine → dry → reconstitute |
Key practical implication: MTBE collection does not require passing the pipette tip through an interphase, which typically reduces matrix carryover. By contrast, chloroform–methanol extraction is more prone to interphase disturbance during collection and may require additional cleanup in challenging matrices.
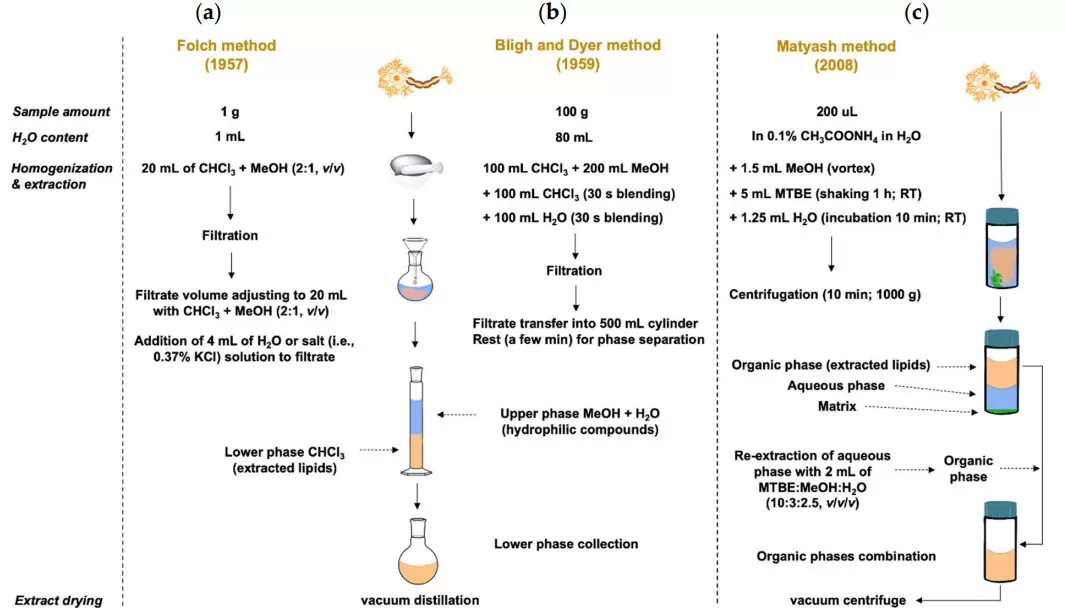
Comparison of lipid extraction methods [3]
Image reproduced from Gerhardtova et al., 2024, Int J Mol Sci, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
3. Strengths and Limitations: Safety, Recovery, and Throughput
When protocols are executed correctly, both approaches can deliver high recoveries for many major lipid classes. Differences are most apparent in operator safety and waste handling, phase separation clarity, automation compatibility, and the recovery of certain polar lipids (notably phosphatidylinositols, PI) in some matrices [3-4].
|
Dimension |
Chloroform–Methanol (Folch/Bligh and Dyer) |
MTBE |
|
Toxicity |
Disadvantage: chloroform is strongly suspected/recognized as carcinogenic; chronic exposure is hazardous. Chloroform can decompose to phosgene and HCl under certain conditions, and may chemically modify labile lipids, potentially affecting analytical accuracy. |
Advantage: low toxicity and non-carcinogenic in typical lab handling; chemically more stable in routine storage/handling and less likely to introduce artifactual modification of unstable lipids, improving operator safety and data integrity. |
|
Extraction efficiency |
Advantage: for many lipid classes (e.g., PE, PC, ceramides), recoveries can reach ~90–98%, historically considered a “gold standard.” Limitation: lower recovery for PI (~68.6%) and greater susceptibility to matrix interference. |
Advantage: comparable recovery for most lipid classes (~90–98%) and substantially higher PI recovery (~91.7%). In plasma comparisons versus Bligh and Dyer, recoveries for some species such as ceramides may be slightly higher. |
|
Phase-separation clarity |
Limitation: interphase can be diffuse; insoluble material may suspend between phases, increasing contamination risk during bottom-phase collection. High density/viscosity of chloroform can make “clean separation” difficult to improve by centrifugation alone. |
Advantage: very clear separation; insoluble material forms a compact bottom pellet, and the boundary between upper MTBE phase and middle aqueous phase is distinct, enabling near-contamination-free collection without extra filtration. |
|
Operational difficulty |
Higher: bottom-phase collection requires careful pipetting control; chloroform volatility requires strict handling conditions; contamination often necessitates additional steps. |
Lower: upper-phase collection is straightforward; lower volatility improves tolerance to minor handling variation; typically no extra filtration, saving time. |
|
Automation / throughput |
Disadvantage: difficult to automate due to bottom-phase aspiration and contamination risk; less suitable for high-throughput workflows. |
Advantage: readily automated using standard liquid-handling robots; well suited for high-throughput lipidomics screening (e.g., large plasma cohorts). |
|
Environmental impact |
Disadvantage: chloroform is a hazardous pollutant; disposal is costly and environmentally burdensome. |
Advantage: comparatively more environmentally friendly and easier to dispose of under standard lab waste-management practices. |
3.1 Additional Technical Considerations (Often Overlooked)
Matrix carryover impacts MS: residual proteins, salts, and detergents can increase ion suppression, contaminate sources, and shorten column lifetime.
Class‑dependent recovery matters: a method that performs well for neutral lipids (TG/CE) may not be optimal for very polar phospholipids.
Solvent purity is non‑negotiable: use LC–MS‑grade solvents and avoid plasticizers (e.g., from soft plastics) that can introduce background ions.
Evaluation of lipid extraction methods for untargeted analysis of mouse tissue lipidome [5]
Image reproduced from Omar et al., 2023, Metabolites, licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0).
4. How to Choose: Best Method by Sample Type and Study Goal
There is no universally best extraction method; the optimal choice depends on matrix complexity, target lipid classes, throughput, and the need for comparability to legacy datasets. Below is a pragmatic starting point aligned with the sample scenarios discussed in the original report.
|
Scenario |
Recommended default |
Why |
|
High-throughput biofluids (plasma/serum/CSF) |
MTBE |
Clean separation; scalable; reproducible |
|
Complex tissues (brain/liver/pancreas) |
MTBE |
Less protein carryover; robust layers |
|
Microbes / whole organisms (E. coli, C. elegans) |
MTBE |
Handles debris; easy batching |
|
Plant tissues (seed/leaf) |
Often CHCl₃–MeOH |
Broad solvation; may improve coverage |
|
Legacy comparability requirement |
CHCl₃–MeOH |
Minimizes method-shift bias |
Notes on Special Cases
- If neutral lipids (TG/CE) are primary endpoints, validate both methods on representative matrices; performance can be sample‑dependent.
- If oxidized or highly labile lipids are targets, minimize heat/light exposure and consider antioxidants (e.g., BHT) and inert gas drying.
- If the sample contains detergents (cell lysis buffers), consider buffer exchange or detergent‑compatible workflows; detergents can severely suppress MS signals.
5. Practical Best Practices to Improve Robustness (Actionable Checklist)
- Add internal standards early: spike isotope‑labeled standards before extraction to quantify recovery and monitor batch drift.
- Control temperature and time: keep samples cold when feasible; standardize incubation/mixing time across all samples.
- Standardize mixing and centrifugation: fix vortex/shake settings, g‑force, duration, and temperature to reduce batch effects.
- Avoid interphase aspiration: if contamination occurs, re‑centrifuge briefly and re‑collect the organic phase.
- Use low‑binding, solvent‑compatible tubes: reduce adsorption and prevent plasticizer contamination.
- Dry gently: avoid excessive heat; use vacuum concentrator or nitrogen; reconstitute in LC‑compatible solvent and fully dissolve (vortex/brief sonication).
- Include blanks and pooled QCs: extraction blanks identify contaminants; pooled QC samples evaluate run stability and CVs.
If you need to switch from chloroform–methanol to MTBE (or vice versa), perform a structured bridging study to avoid introducing systematic bias: i) Extract the same pooled matrix with both methods (≥5 replicates each) and analyze under identical LC–MS conditions; ii) Compare class‑level totals and species‑level profiles; evaluate recovery using internal standards and assess coefficients of variation (CVs); iii) Inspect ion suppression indicators (e.g., internal‑standard response) and background contaminants (from blanks); iv) If systematic shifts are observed, document method‑specific baselines and avoid mixing methods within a single cohort when possible.
6. Summary: Legacy Gold Standard vs. Modern High-Throughput Workflow
As a long-standing “gold standard” for over half a century, chloroform–methanol extraction remains useful for conventional applications and relatively simple matrices. However, its inherent drawbacks—high toxicity, operational complexity, and limited compatibility with automation—have led to its gradual replacement by MTBE-based protocols in many workflows.
MTBE extraction offers lower toxicity, high and consistent recovery, clearer phase separation, and strong suitability for automation. Overall, it delivers performance comparable to chloroform–methanol extraction and can provide superior recovery for certain lipid classes, notably phosphatidylinositols (PI). These advantages make it particularly well aligned with contemporary lipidomics needs, including high-throughput processing and complex biological matrices, such as clinical screening and lipid profiling of challenging tissues.
Regardless of the method selected, strict adherence to key parameters—solvent ratios, incubation time, and centrifugation conditions—is essential to ensure stability and reproducibility. Because extraction performance directly shapes downstream analytical quality, strict protocol adherence is a primary determinant of overall study success.
References
1. FOLCH J, LEES M, SLOANE STANLEY GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957 May;226(1):497-509.
2. Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008 May;49(5):1137-46. doi: 10.1194/jlr.D700041-JLR200.
3. Gerhardtova I, Jankech T, Majerova P, Piestansky J, Olesova D, Kovac A, Jampilek J. Recent Analytical Methodologies in Lipid Analysis. Int J Mol Sci. 2024 Feb 13;25(4):2249. doi: 10.3390/ijms25042249.
4. Reis A, Rudnitskaya A, Blackburn GJ, Mohd Fauzi N, Pitt AR, Spickett CM. A comparison of five lipid extraction solvent systems for lipidomic studies of human LDL. J Lipid Res. 2013 Jul;54(7):1812-24. doi: 10.1194/jlr.M034330.
5. Omar AM, Zhang Q. Evaluation of Lipid Extraction Protocols for Untargeted Analysis of Mouse Tissue Lipidome. Metabolites. 2023 Sep 9;13(9):1002. doi: 10.3390/metabo13091002.
Read more
- Fatty acids: its classification, synthesis pathways and related diseases
- A Complete Analysis of Saturated Fatty Acids: from Food Sources to Health Effects
- Sterol Lipids: Structure, Function, and Their Role in Health and Disease
- A Comprehensive Guide to Quantitative Lipidomics: Methodologies, Workflows, and Applications
- LC-MS vs GC-MS in Lipidomics: How to Choose the Best Method for Lipid Analysis
- LIPID MAPS Lipid Classification and Nomenclature Rules: A Practical Guide
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


