LIPID MAPS Lipid Classification and Nomenclature Rules: A Practical Guide
When people hear the term “lipids,” many instinctively think of “fat” or “oil.” In fact, lipids comprise a large and highly diverse class of biomolecules that are ubiquitous in plant and animal cells and play indispensable roles in energy storage, membrane architecture, and signal transduction. Given the vast number of lipid species and their pronounced structural heterogeneity, a standardized framework for lipid classification and nomenclature is essential. This report provides a detailed overview of the most widely recognized lipid classification system—the ‘LIPID MAPS’ framework—highlights key structural features across major lipid categories using representative examples, and summarizes fundamental nomenclature principles to facilitate accurate interpretation of lipid identities [1-2].
Why a Unified Lipid Classification and Naming System Matters?
From a chemical perspective, many lipids are esters formed between fatty acids and alcohols, along with related derivatives. A defining physicochemical characteristic of lipids is their poor solubility in water and high solubility in nonpolar organic solvents (e.g., diethyl ether and chloroform). Meanwhile, lipids display exceptional structural diversity—ranging from simple triacylglycerols to complex sphingolipids and sterols—where variations in core scaffolds and substituents can be substantial.
Prior to the establishment of the LIPID MAPS framework, lipid classification often relied on superficial properties such as hydrolysis behavior and solubility, without a robust and systematic structural basis. Consequently, the same lipid could be described by multiple names, and the boundaries between lipid classes were frequently ambiguous, which significantly impeded communication and progress in lipid research. In 2005, the ‘LIPID MAPS Consortium’ introduced a comprehensive classification system grounded in lipid chemical structures and biosynthetic relationships, enabling standardized and systematic lipid taxonomy and becoming a widely adopted global reference in lipidomics.
LIPID MAPS Classification: The 8 Major Lipid Categories (with Examples)
The LIPID MAPS system classifies all lipids into ‘eight major categories’ [3]. Each category is further partitioned into subclasses and sub-subclasses based on structural criteria, forming a clearly defined hierarchical taxonomy. Each lipid is assigned a unique ‘LIPID MAPS ID (LM_ID’ to support retrieval and unambiguous identification. Among these, the first seven categories represent the most commonly encountered core lipid classes. Below, we summarize these classes and describe their key structural features with representative examples.
(1) Fatty Acids (FA): Core Structure and Key Terms
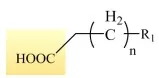
Fatty acids are fundamental structural units of lipids; nearly all complex lipids incorporate fatty acids as building blocks. Structurally, fatty acids consist of a long hydrocarbon (alkyl) chain and a terminal carboxyl group (-COOH). Chain lengths typically range from ‘4 to 24 carbons’ and fatty acids are broadly categorized as ‘saturated’ or ‘unsaturated’.
Core structure (stearic acid as a representative saturated fatty acid): CH3(CH2)16COOH
Structural details: Stearic acid contains an 18-carbon hydrocarbon chain in which all carbon–carbon bonds are single bonds (i.e., fully saturated). The chain is relatively linear and packs efficiently, contributing to a high melting point (~70°C) and a solid physical state at room temperature.
Unsaturated fatty acid (oleic acid as a representative example): CH3(CH2)7CH = CH(CH2)7COOH
Structural details: Oleic acid is an 18-carbon fatty acid with a single double bond between carbons 9 and 10. The double bond—typically in the cis configuration in biological systems—introduces a conformational “kink,” reduces packing efficiency, lowers the melting point (~13°C), and results in a liquid state at room temperature. Unsaturated fatty acids can be further classified as monounsaturated (one double bond) or polyunsaturated (two or more double bonds). Polyunsaturated fatty acids such as linoleic acid and α-linolenic acid are considered essential in humans because they cannot be synthesized endogenously and must be obtained through diet.
Within LIPID MAPS, fatty acids are further categorized according to chain length and the number and positions of double bonds, serving as a foundational layer for lipid classification.
(2) Glycerolipids (GL): Triacylglycerols and Energy Storage
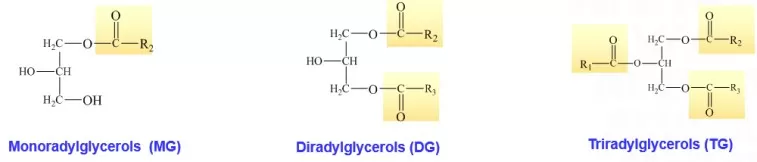
Glycerolipids structure
Glycerolipids use glycerol (propane-1,2,3-triol) as the backbone. The three hydroxyl groups (-OH) of glycerol undergo esterification with fatty acids to form ester linkages (-COO-). Glycerolipids represent the most abundant lipid class in nature (e.g., the principal constituents of animal and plant fats and oils), with triacylglycerols (triglycerides) as the canonical representatives.
Core structure of triacylglycerol: CH2OCOR1 - CHOCOR2 - CH2OCOR3 (R₁, R₂, R₃ denote fatty acyl chains that may be identical or different)
Structural details: Each of the three glycerol carbons is esterified to a fatty acid. When all three fatty acyl chains are identical (e.g., tristearin), the molecule is termed a “simple” triacylglycerol; when the chains differ (e.g., stearoyl–oleoyl–palmitoyl glycerol), it is a “mixed” triacylglycerol. Triacylglycerols are the primary energy storage form in organisms; oxidation of 1 g of triacylglycerol releases approximately 37 kJ of energy, exceeding that of carbohydrates and proteins. They also contribute to thermal insulation and mechanical protection.
In addition to triacylglycerols, glycerolipids include monoacylglycerols (one acyl chain) and diacylglycerols (two acyl chains), both of which serve as important intermediates in lipid metabolism and signaling.
(3) Glycerophospholipids (GP): Membrane Lipids and Head Groups
Glycerophospholipids also use glycerol as the backbone but are structurally more complex. Specifically, the hydroxyl groups at the sn-1 and sn-2 positions are esterified with fatty acids, whereas the sn-3 hydroxyl group is linked to a phosphate group (-PO₄³⁻). The phosphate group is further connected to a polar head group (e.g., choline or ethanolamine). As a result, glycerophospholipids are amphipathic, featuring hydrophobic fatty acyl tails and a hydrophilic phosphate-containing head, and constitute core structural components of biological membranes via lipid bilayer formation.
Representative structure—Phosphatidylcholine (lecithin): CH2OCOR1 - CHOCOR2 - CH2OPO3- - OCH2CH2N+(CH3)3
Structural details: The sn-1 and sn-2 positions are esterified to fatty acyl chains (commonly, sn-1 is saturated and sn-2 is unsaturated). The sn-3 position is linked to choline through a phosphodiester bond, forming the polar head group. This amphipathic organization promotes spontaneous bilayer assembly in aqueous environments, with hydrophilic heads oriented toward the aqueous phase and hydrophobic tails packed inward, thereby establishing the fundamental framework of cell membranes. Beyond structural roles, glycerophospholipids participate in signaling and molecular transport. Common glycerophospholipids also include phosphatidylethanolamine and phosphatidylserine, which are widely distributed in egg yolk, soybeans, brain tissue, and other biological sources.
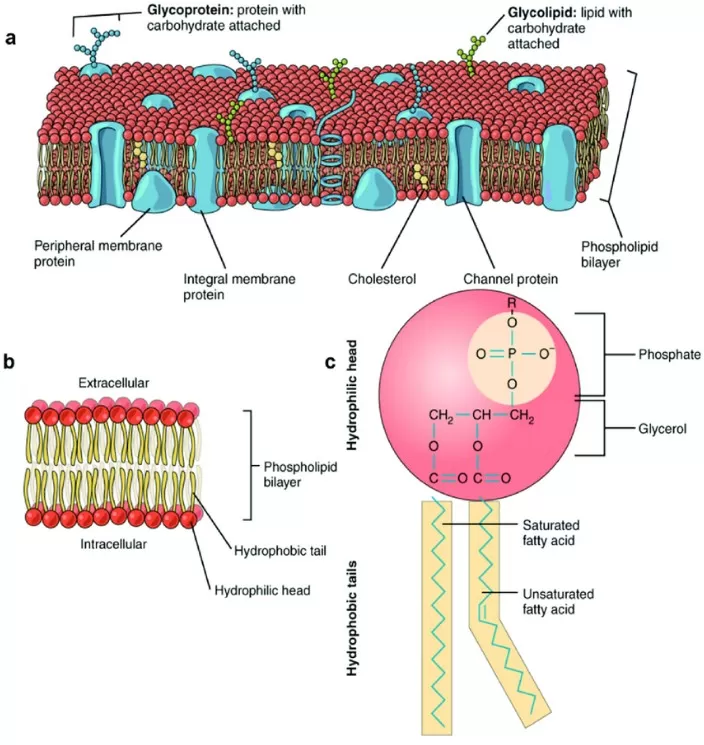
Cell membrane and glycerophospholipids
Common head-group relationships (X + phosphatidic acid → corresponding phospholipid):
- No X substituent → phosphatidic acid (PA)
- Choline + phosphatidic acid → phosphatidylcholine (PC)
- Serine + phosphatidic acid → phosphatidylserine (PS)
- Inositol + phosphatidic acid → phosphatidylinositol (PI)
- Ethanolamine + phosphatidic acid → phosphatidylethanolamine (PE)
- Glycerol + phosphatidic acid → phosphatidylglycerol (PG)
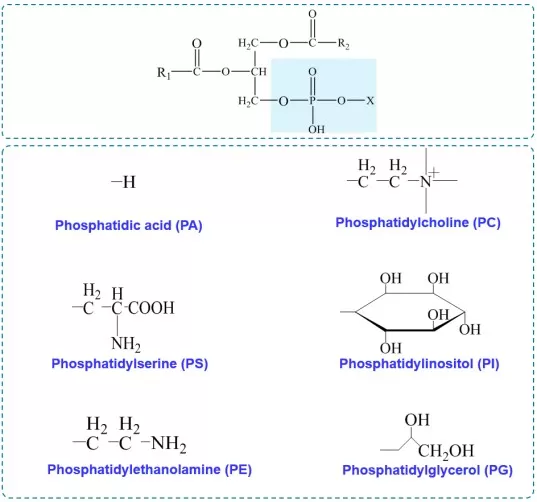
Glycerophospholipids structure
(4) Sphingolipids (SP): Ceramides, Sphingomyelins, and Glycosphingolipids
In contrast to glycerol-based lipids, sphingolipids are built on a sphingoid base (sphingosine) rather than glycerol. Sphingosine contains an amino group (-NH₂), a hydroxyl group (-OH), and a long hydrocarbon chain. The amino group forms an amide bond with a fatty acid, and the hydroxyl group can be further substituted with a polar head group (e.g., phosphate or carbohydrates). Sphingolipids are likewise amphipathic and are enriched in animal cell membranes, particularly in nervous tissue.
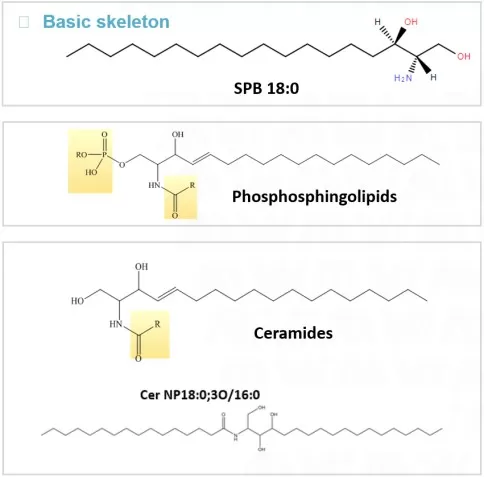
Sphingolipids structure
Representative structure—Ceramide (the core scaffold of sphingolipids): CH3(CH2)12CH = CHCH(OH)CH(NHCOCH2(CH2)nCH3)CH2OH
Structural details: Ceramide consists of sphingosine linked to a fatty acid via an amide bond and serves as the precursor for sphingomyelins and glycosphingolipids. When the ceramide hydroxyl group is substituted with phosphocholine, it forms sphingomyelin, a major constituent of the myelin sheath. When substituted with sugar residues (e.g., glucose or galactose), it forms glycosphingolipids, which participate in cell recognition and neural signaling.
(5) Sterols (ST): Cholesterol and Sterol Biology
Sterols are structurally distinct from the lipid classes described above. Their core scaffold is cyclopentanoperhydrophenanthrene, comprising four fused rings (three six-membered rings and one five-membered ring). Substituents such as methyl groups, hydroxyl groups, and alkyl side chains confer structural diversity and give rise to a broad class of biologically important derivatives.
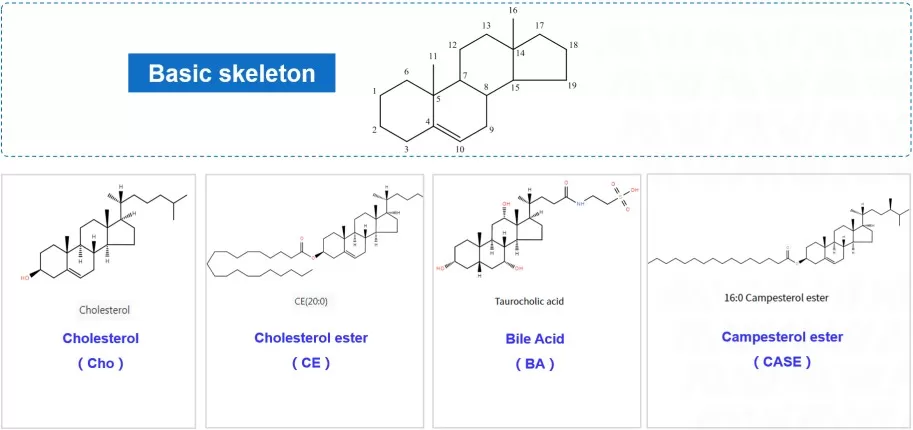
Sterols structure
Representative structure—Cholesterol: C27H46O (A/B/C fused six-membered rings + D five-membered ring; hydroxyl at C3; eight-carbon alkyl chain at C17)
Structural details: Cholesterol is a major component of animal cell membranes and modulates membrane fluidity, helping prevent excessive rigidification at low temperatures and excessive fluidity at high temperatures. It also serves as a biosynthetic precursor of bile acids, steroid hormones (e.g., testosterone and estradiol), and vitamin D. Plants do not contain cholesterol; instead, they contain phytosterols (e.g., β-sitosterol), which are structurally similar and can reduce intestinal cholesterol absorption in humans. Importantly, cholesterol lacks ester bonds and is therefore classified as a non-saponifiable lipid, meaning alkaline hydrolysis does not generate soaps.
(6) Prenol Lipids (PR): Isoprene-Derived Lipids
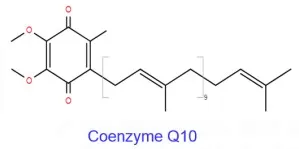
Prenol lipids are derived from the condensation of isoprene units (i.e., terpenes/terpenoids) and encompass a broad range of structurally diverse molecules, for which systematic nomenclature can be challenging. Their repeating C₅H₈ units are connected through carbon–carbon bonds, and their oxygenated derivatives and compounds with varying degrees of saturation are typically hydrophobic. Notable prenol lipids include quinones and phenolic compounds such as vitamin E, vitamin K, and coenzyme Q10 (ubiquinone-10).
(7) Glycolipids (GLy): Sugar-Containing Membrane Lipids
Glycolipids may employ either glycerol or sphingosine as the backbone. Their defining characteristic is that backbone hydroxyl group(s) are linked to carbohydrate moieties (mono-, di-, or polysaccharides) via glycosidic bonds rather than phosphate groups. Glycolipids are amphipathic and are predominantly localized to the outer leaflet of cell membranes.
Representative structure—Cerebroside (a glycosphingolipid): Sphingosine-fatty acid amide bond-glucose/galactose
Structural details: Cerebrosides are sphingosine-based lipids in which, after amide bond formation with a fatty acid, the hydroxyl group is glycosylated with a single sugar (glucose or galactose). They are abundant in neural membranes and contribute to neural function. More complex glycolipids, such as gangliosides, contain multiple sugars and sialic acid residues and play important roles in cell recognition and immune-related processes. In plants, glycerol-based glycolipids (glyceroglycolipids) are also common; they comprise glycerol linked to fatty acids and sugars and are major lipid constituents of chloroplast membranes.
Core Lipid Nomenclature Rules (LIPID MAPS-Aligned)
Lipid nomenclature includes both systematic nomenclature (IUPAC-based; rigorous and standardized) and common nomenclature (concise and widely used in practice). The LIPID MAPS framework supports both approaches, with naming conventions centered on accurate description of structural composition [1,2]. Below, we summarize nomenclature rules for three frequently encountered lipid types.
(1) Naming Rules for Fatty Acids
The central convention is “carbon chain length + number of double bonds + double-bond positions,” typically expressed as: C:D Δ(position[s])
where C denotes the number of carbon atoms, D denotes the number of double bonds, and Δ specifies the position(s) of double bonds counted from the carboxyl terminus.
Examples:
- Stearic acid: 18 carbons, 0 double bonds → systematic name “octadecanoic acid”; shorthand “18:0” (Δ notation is unnecessary in the absence of double bonds).
- Oleic acid: 18 carbons, 1 double bond at C9–C10 (with the carboxyl carbon designated as C1) → systematic name “9-octadecenoic acid”; shorthand “18:1Δ⁹”.
- Linoleic acid: 18 carbons, 2 double bonds at Δ9 and Δ12 → shorthand “18:2Δ9,12”. It also belongs to the ω-6 series (counting from the methyl terminus, the first double bond is at the 6th carbon) and can be written as “18:2ω-6”.
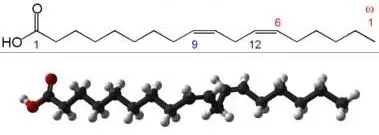
Linoleic acid structure
(2) Naming Rules for Triacylglycerols (Triglycerides)
Triacylglycerol nomenclature is based on fatty acyl composition on the glycerol backbone. Common naming typically lists the three fatty acids (often corresponding to positions 1, 2, and 3), whereas systematic nomenclature explicitly specifies stereospecific positions.
Examples:
- Tristearin: three stearic acid chains → common name “tristearin”; systematic name “1,2,3-tristearoyl-sn-glycerol,” where sn- denotes stereospecific numbering and LIPID MAPS standardizes the use of sn-configuration.
- Mixed triacylglycerol (1-stearoyl-2-oleoyl-3-palmitoyl glycerol): sn-1 is stearic acid, sn-2 is oleic acid, and sn-3 is palmitic acid → often simplified as “stearoyl–oleoyl–palmitoyl glycerol.”
(3) Naming Rules for Glycerophospholipids
The core convention is “head group + fatty acyl composition + glycerol backbone,” commonly expressed as: phosphatidyl-(head group) (FA1/FA2)
When available, the stereospecific positions and structural details of fatty acids should be specified.
Examples:
Phosphatidylcholine (lecithin): head group is choline; sn-1 is stearic acid (18:0) and sn-2 is oleic acid (18:1Δ⁹) → “1-stearoyl-2-oleoyl-phosphatidylcholine,” abbreviated as “POPC” (acyl shorthand: P = palmitic acid, O = oleic acid, S = stearic acid).
Phosphatidylethanolamine: head group is ethanolamine with the same acyl composition → “1-stearoyl-2-oleoyl-phosphatidylethanolamine,” abbreviated as “POPE”.
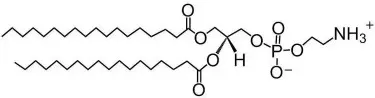
Phosphatidylethanolamine structure
Summary: How to Read Lipid Names with Confidence
The primary value of the LIPID MAPS classification system lies in its structure-driven, function-aware organization. It partitions chemically diverse lipids into eight major classes, assigns each lipid a unique identifier (LM_ID), and resolves inconsistencies and ambiguities inherent to earlier classification approaches. At its core, lipid nomenclature provides a structured way to encode molecular composition: whether describing the chain length and unsaturation pattern of fatty acids or specifying the backbone and substituents of glycerolipids and phospholipids, proficiency in recognizing structural features enables efficient interpretation of lipid names.
As essential biomolecules in living systems, lipids underpin fundamental research across biology, medicine, and related disciplines [4]. Even in everyday contexts such as nutrition and health, the ability to interpret lipid structures and names supports a clearer understanding of lipid function and biological significance.
References
1. Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, Merrill AH Jr, Murphy RC, Raetz CR, Russell DW, Subramaniam S. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007 Jan;35(Database issue):D527-32. doi: 10.1093/nar/gkl838.
2. Conroy MJ, Andrews RM, Andrews S, Cockayne L, Dennis EA, Fahy E, Gaud C, Griffiths WJ, Jukes G, Kolchin M, Mendivelso K, Lopez-Clavijo AF, Ready C, Subramaniam S, O'Donnell VB. LIPID MAPS: update to databases and tools for the lipidomics community. Nucleic Acids Res. 2024 Jan 5;52(D1):D1677-D1682. doi: 10.1093/nar/gkad896.
3. Liebisch G, Fahy E, Aoki J, Dennis EA, Durand T, Ejsing CS, Fedorova M, Feussner I, Griffiths WJ, Köfeler H, Merrill AH Jr, Murphy RC, O'Donnell VB, Oskolkova O, Subramaniam S, Wakelam MJO, Spener F. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J Lipid Res. 2020 Dec;61(12):1539-1555. doi: 10.1194/jlr.S120001025.
4. Tsugawa H, Ikeda K, Takahashi M, Satoh A, Mori Y, Uchino H, Okahashi N, Yamada Y, Tada I, Bonini P, Higashi Y, Okazaki Y, Zhou Z, Zhu ZJ, Koelmel J, Cajka T, Fiehn O, Saito K, Arita M, Arita M. A lipidome atlas in MS-DIAL 4. Nat Biotechnol. 2020 Oct;38(10):1159-1163. doi: 10.1038/s41587-020-0531-2.
Read more
- Fatty acids: its classification, synthesis pathways and related diseases
- A Complete Analysis of Saturated Fatty Acids: from Food Sources to Health Effects
- Sterol Lipids: Structure, Function, and Their Role in Health and Disease
- A Comprehensive Guide to Quantitative Lipidomics: Methodologies, Workflows, and Applications
- LC-MS vs GC-MS in Lipidomics: How to Choose the Best Method for Lipid Analysis
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


